Abstract
The Human BioMolecular Atlas Program (HuBMAP) provides an opportunity to contextualize findings across cellular to organ systems levels. Constructing an atlas target is the primary endpoint for generalizing anatomical information across scales and populations. An initial target of HuBMAP is the kidney organ and arterial phase contrast-enhanced computed tomography (CT) provides distinctive appearance and anatomical context on the internal substructure of kidney organs such as renal context, medulla, and pelvicalyceal system. With the confounding effects of demographics and morphological characteristics of the kidney across large-scale imaging surveys, substantial variation is demonstrated with the internal substructure morphometry and the intensity contrast due to the variance of imaging protocols. Such variability increases the level of difficulty to localize the anatomical features of the kidney substructure in a well-defined spatial reference for clinical analysis. In order to stabilize the localization of kidney substructures in the context of this variability, we propose a high-resolution CT kidney substructure atlas template. Briefly, we introduce a deep learning preprocessing technique to extract the volumetric interest of the abdominal regions and further perform a deep supervised registration pipeline to stably adapt the anatomical context of the kidney internal substructure. To generate and evaluate the atlas template, arterial phase CT scans of 500 control subjects are de-identified and registered to the atlas template with a complete end-to-end pipeline. With stable registration to the abdominal wall and kidney organs, the internal substructure of both left and right kidneys are substantially localized in the high-resolution atlas space. The atlas average template successfully demonstrated the contextual details of the internal structure and was applicable to generalize the morphological variation of internal substructure across patients.
Keywords: Computed Tomography, Kidney Substructure Atlas, Abdomen Atlas, Medical Image Registration
1. INTRODUCTION
Considerable effort has been made by the Human BioMolecular Atlas Program (HuBMAP) to relate molecular findings in organ anatomy across cellular to organ systems levels [1]. With the previous efforts of mapping the organization and molecular to profile cells across different tissues and organs [2], it is vitally important to contextualize the anatomical characteristics of organs in well-defined reference templates to act as a common framework for contextualizing multimodal molecular information across scales. Computed tomography (CT) provides an imaging platform to visualize anatomical context at organ system level. Contrast enhancement, is used to emphasize the structural and anatomical context between neighboring organs with the injection of contrast agent and guide to extract information from regions of interest (ROI) [3]. The kidney is the initial target of HuBMAP to analyze the structural anatomy across scales. Arterial phase CT provides distinctive representation for further analysis of the kidney substructure perspective [4]. Figure 1 illustrates how the kidneys are localized with significant variability of size and anatomical regions. Additionally, the surface rendering of each internal structure demonstrates the large volumetric difference across patients from different demographics. It is challenging to adapt such variable morphological characteristics in a single well-defined reference for population analysis.
Figure 1:
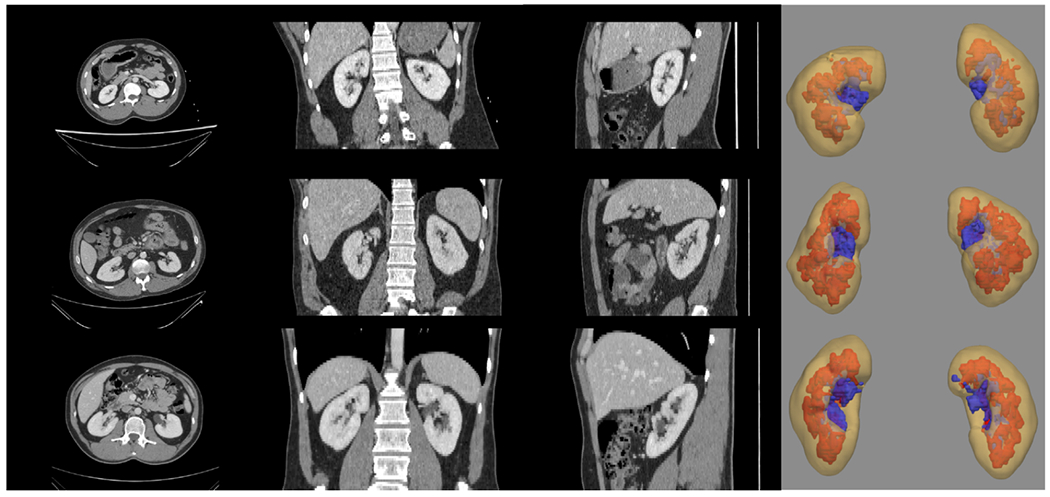
Significant variations in contrast intensity and morphology are demonstrated in both the external and internal structure of the kidney (1) yellow: renal cortex, 2) red: medulla, and 3) pelvicalyceal system). The asymmetric property in the kidney appearance and the anatomical (such as size and position) variation is shown and it is challenging to adapt a well-defined anatomical reference for kidney organs.
Here, we aim to adapt the conventional information of each kidney substructure on a single anatomical atlas template. Previous works have been demonstrated in building an atlas platform with neuroimaging [5, 6]. Multiple brain atlases are built to reveal the population characteristics of brains in both adults and infants [7–9]. Apart from looking into the anatomical characteristics, atlas reference is used as a platform to perform segmentation with unsupervised settings [10, 11]. Multiple atlas references are randomly picked and perform registration between the subject moving scans to the multiple atlases’ platform [12]. Segmentation predictions are computed with joint label fusion using the guidance of multiple registered outputs. However, there has been more limited work in generating atlas frameworks for the specific organs in abdominal regions [13, 14]. With the variability of demographics, several challenges are raised with 1) the difference of abdominal body shape and 2) the performance of registrations for adapting large deformation. Currently, registration pipelines are specifically designed for the abdominal regions to increase the robustness of adapting the abdominal organ-corresponding information such as DEEDS [15–17]. With the DEEDS registration pipeline, over-deformation is demonstrated and leads to great variance in registering the liver and spleen regions due to the field of view varies across patients with different imaging protocols [13, 14]. Therefore, a stable atlas construction pipeline is needed to generalize the organ context in the abdomen across demographics and construct a well-defined atlas target with a high successful rate of transferring ROI context needed for anatomical evaluation.
In this study, we construct a contrast-preserving kidney substructure atlas with a deep unsupervised preprocessing and registration pipeline to increase the robustness of adapting organ context across patients in high resolution. With a total of 500 arterial subject scans with healthy kidneys, we generated the average mapping of healthy kidney substructures across this population. Atlas target and moving subject scans are initially downsampled to input for the deep registration pipeline. The predicted displacement field is upsampled back to the atlas resolution and warped with the upsampled subject images. Registration performances are evaluated in both quantitative and qualitative perspectives and the kidney substructure context is stably transferred with contrastive and morphological characteristics to the atlas space.
2. METHODS
2.1. Preprocessing
With demographic variability across patients, the field of view varies significantly in 3D abdominal CT scans across populations and may contribute to the possibility of registration failure. Here, we introduce a deep learning preprocessing pipeline body part regression to generate anatomical information for cropping the kidney-corresponding area of interest to perform stable registration [18, 19]. Specifically, 2D axial slices are initially extracted from each scan and a prediction score is generated to identify the approximate anatomical location in the human body for each slice. The output value for each slice is in the range of −12 to + 12 (arbitrary units), which specify regions from the heart to the pelvis. We defined the value between −4 to 3 as the kidney regions of interest and crop the volumetric scans to ensure a similar field of view between atlas target and subject scans.
2.2. Registration Pipeline
To adapt to the significant variation of the abdominal regions across patients, a deep network registration pipeline is introduced with probability dense displacement networks (PDD-Net), which aims to align a 3D moving image Im to the fixed image If space with an optimal spatial transformation learned on the extracted deep feature context [20, 21]. The complete overview of the registration pipeline is illustrated in Figure 2. Both preprocessed subject scans and atlas target are input into a small network to learn a meaningful non-linear mapping to align from input intensity to a dense feature context. The Obelisk approach is employed for the small network f to effectively capture the spatial context with significant deformation and a normal 5 × 5 × 5 convolution kernel is added to learn the edge feature for body-aligned registration [21]. After the extraction of the spatial feature context, our goal is to predict an optimized displacement field δ(k) ← ν that apply a vector ν to every set of control points on a grid for non-linear transformation and additionally achieve the best similarity between the organ labels. With the use of conventional discrete registration [15] and the correlation layer in [22], we sample the context from discrete grids and compute a 6D tensor D representing the dissimilarities with the feature dimension z. Here, we use the negated mean square error across the feature dimension to generate the 6D tensor:
| (1) |
Figure 2:
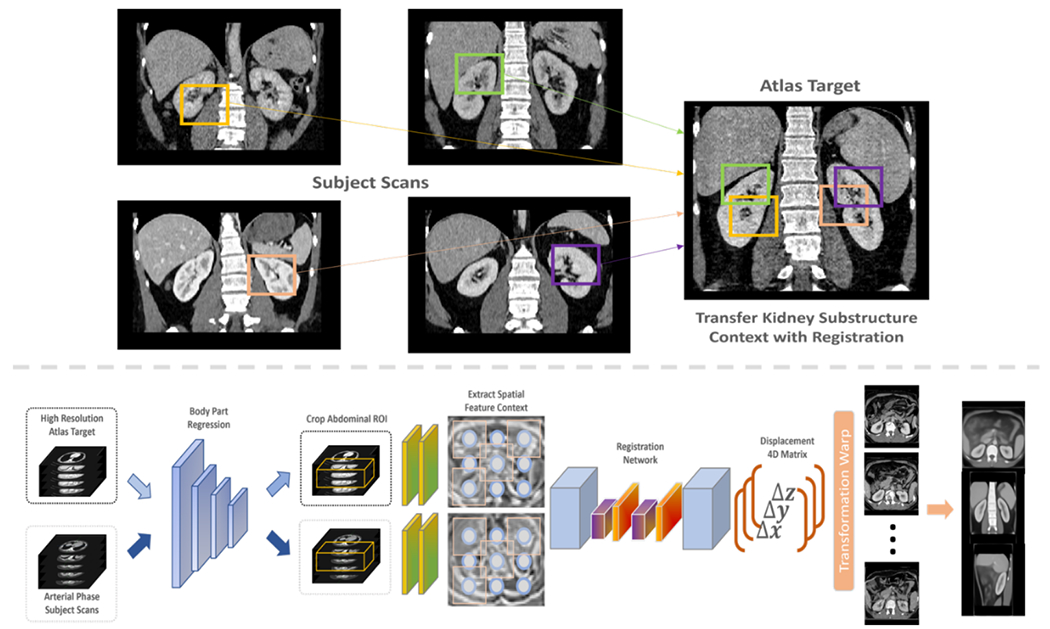
We aim to transfer the significant variability of contrast and morphology to a healthy-defined anatomical atlas target with linear and non-linear transformations for generalizing anatomical context across scales (top panel). The complete pipeline (lower panel) can be divided into two steps: 1) body part regression preprocessing and 2) deep supervised registration. We initially crop the abdominal area of interest for both atlas target and subject scans with the guidance of body part regression network. We downsample both volumes and input into a deep registration network to predict the voxel displacement across tri-planar perspective. We finally warp the predicted transformations to each subject scan and compute average map for analysis.
As abdominal structures vary significantly across demographics, it causes over-deformation and the registered images to become ill-posed with non-linear registration. PDD-Net provides an opportunity to model the regularization constraints with diffusion regularization and use fast mean-field inference with two iterations only for discrete optimization [23]. It consists of two steps: 1) transformation with label compatible that assigns on spatial control points and 2) the additional average pooling layers with stride 1 for filtering message. Previous work demonstrated that the diffusion regularization can be generated using min-convolutions with a lower envelope of parabolas rooted at the offset of 3D displacement with heights equivalent to the sum of dissimilarity terms and the previous iteration of mean-field inference [24]. We approximately compute the diffusion regularization for dense displacement with a min-pooling layer to extract the local minima in the cost tensor. Two average pooling layers are additionally used for smoothing the context extracted. The integration of min-pooling and average pooling aims to perform the regularization approach in multiple dimensions: 1) 3 displacement dimensions (min-convolution) and 2) 3 spatial dimensions (mean-field inference) for end-to-end optimization.
2.3. Deep Label Supervision for Registration
Apart from adapting the dissimilarities in intensity level, we aim to adapt the anatomical context of a specific region of interest (ROI) with high stability. Here, we use a supervised label loss term to preserve the morphological information of ROI instead of over-deformation. We use a SoftMax activation to compute a probability mapping with the regularized output over the displacement context. One-hot representations from the moving subject segmentations are wrapped in the corresponding spatial location with predicted displacements and compute the dissimilarity between wrapped subject labels and fixed target labels with mean square error (MSE). The predicted displacement is a 4D multi-channel output and the number of channels represented the displacement field on each direction (Δx, Δy, Δz). The final prediction is resampled to the original input resolution with tri-linear interpolation.
3. DATA AND EXPERIMENTS
3.1. Data and Platform
To evaluate the atlas template, abdomen CT volumetric scans from 1971 patients were retrieved in de-identified form from ImageVU with the approval of the Institutional Review Board (IRB) (IRB number: 160764). Exclusion criteria are set based on ICD-9 codes to include subjects with healthy kidney organs only. Out of 1971, 500 subjects are retrieved with the assurance of ICD-9 codes and within the age range of 18 to 50 years old. We limited our studies to the subjects with arterial phase CT only, as only the arterial phase CT provides adequate data on the distinctive anatomy of kidney substructures. Therefore, in a total of 500 subjects 3D volumetric CT are used to generate and evaluate the atlas template. For the atlas target image, a single subject volume with high resolution (0.8 × 0.8 × 0.8) is used with a dimension of 512 × 512 × 434. The criteria to choose data as the atlas template are based on the contrastive and morphological characteristics of the kidneys. With the use of body part regression preprocessing, both subject scans and atlas target are cropped to the abdominal interest only. We downsampled the subject scans and atlas target to an isotropic voxel resolution of 1.5mm × 1.5mm × 1.5mm and a dimension of 192 × 160 × 256. The downsampled scans are then input into the deep registration pipeline to predict the displacement field for transferring the kidney substructure context to the atlas space.
3.2. Experiments
3.2.1. Registration Comparison
We performed a conventional registration algorithm DEnsE Displacement Sampling (DEEDS) as our baseline method and previous works have demonstrated that DEEDS outperformed other traditional registration tools (ANTS, NiftyReg) in the inter-patient 3D abdominal CT registration study. DEEDS calculate the image similarity between a number of random sampling voxels of each control point and model the diffusion regularization through a pair-wise term of the displacement field. Hierarchical steps are performed following with 1) DEEDS affine registration and 2) DEEDS deformable registration. A displacement matrix is an output to provide the transformation information corresponding to each control point. We apply the displacement matrix to the subject ground truth labels and compare the transformed label with the atlas target label for evaluating the registration performance. The ground truth label of kidney substructure corresponds to the renal cortex (label: 1), medulla (label: 2), and pelvicalyceal system (label: 3) [4]. Dice Coefficient is used to measure the overlapping regions between the prediction label and ground truth labels.
| (2) |
3.2.2. Atlas Construction
Both subject scans and atlas target are input corresponding to the moving image and fixed image respectively. As the image inputs have to be downsampled to fit into the deep registration pipeline without memory outage, we aim to minimize the loss of the high-resolution context from the atlas target. After we output the predicted displacement field, we further resample the displacement field to the original resolution of the atlas target with tri-linear interpolation and warp the displacement field on upsampled subject scans. All warped upsampled scans are summed and generate the average map to evaluate the transition of the anatomical context across kidney substructures qualitatively.
3.2.3. Deep Registration Model
We initially use the pre-trained model from [20], which trained on 10 contrast-enhanced 3D CT scans of the VISERAL3 training dataset. Each scan in VISERAL3 training dataset consists of nine anatomical structures well-annotated including 1) liver, 2) spleen, 3) pancreas, 4) gallbladder, 5) urinary bladder, 6) right kidney, 7) left kidney, 8) right psoas major muscle and 9) left psoas major muscle [25]. All images are downsampled to an isotropic voxel of 2.0mm × 2.0mm × 2.0mm with dimensions of 192 × 160 × 256 for training input and no initial affine alignment is performed between the subject scans and fixed target scan.
4. RESULTS
To evaluate the generalizability of the atlas template on the localizing contextual information of kidney substructures, we first compare the single subject registration qualitatively, and manually visualize the quality of the registered kidney substructures. As shown in Figure 4, the registered output from DEEDS demonstrates a fair registration quality of transferring both left and right kidney information. However, over-deformation is demonstrated with a small extent to the left kidney structure and the boundary information between the right kidney and liver cannot be clearly separated. With the PDD-Net registration, the context of the left and right kidney is comparable to the anatomical information of the kidneys in the atlas target. The substructure context is stably transferred to the atlas-defined space with limited over-deformation. We further evaluate the atlas target across all populations and generate the average mapping to visualize the localizing context of the kidney substructure in the atlas template. The average mapping generates with PDD-Net demonstrates a more distinctive appearance in the kidney substructures, while a certain extent of blurriness in the kidney substructure context is shown with DEEDS registration. The contrastive and morphological characteristics of the kidney substructures are transferred with stability in the atlas target across all populations. In terms of label-wise measure, we apply the predicted displacement field to the moving subject labels and compare the similarity between the warped labels and the atlas labels in Figure 3. Significant improvement (p < 0.001) on the renal cortex and pelvicalyceal system segmentation is demonstrated with PDD-Net, while DEEDS demonstrates a better performance on the medulla segmentation. The medulla segmentation with PDD-Net demonstrates a significant decrease of variance in performance and shows the generalizability across the hard registration case for kidneys.
Figure 4:
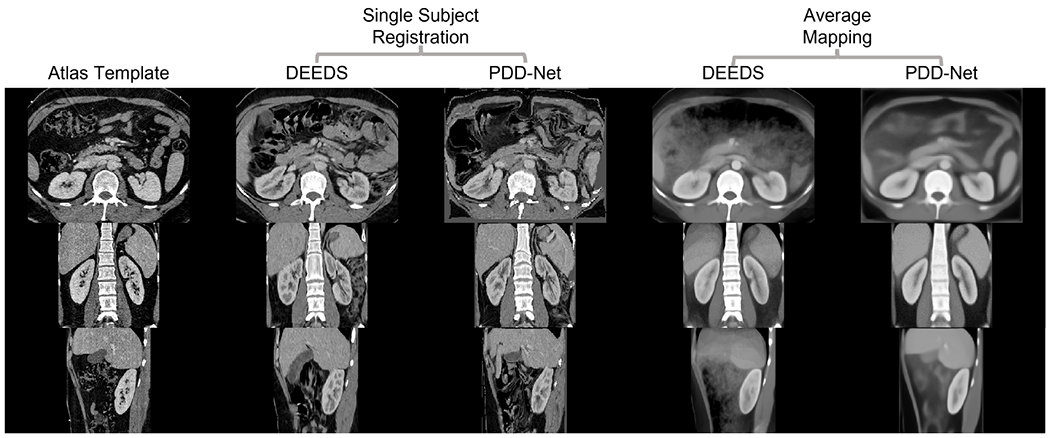
This qualitative representation demonstrates the comparison of transferring kidney substructure information with DEEDS and PDD-Net. The single subject is arbitrary picked and shows that the renal structure is comparatively over-deformed with DEEDS. From the average template, the contrastive and morphological context of the substructure is illustrated more appealing both in the boundary and the cortex anatomy with PDD-Net.
Figure 3:
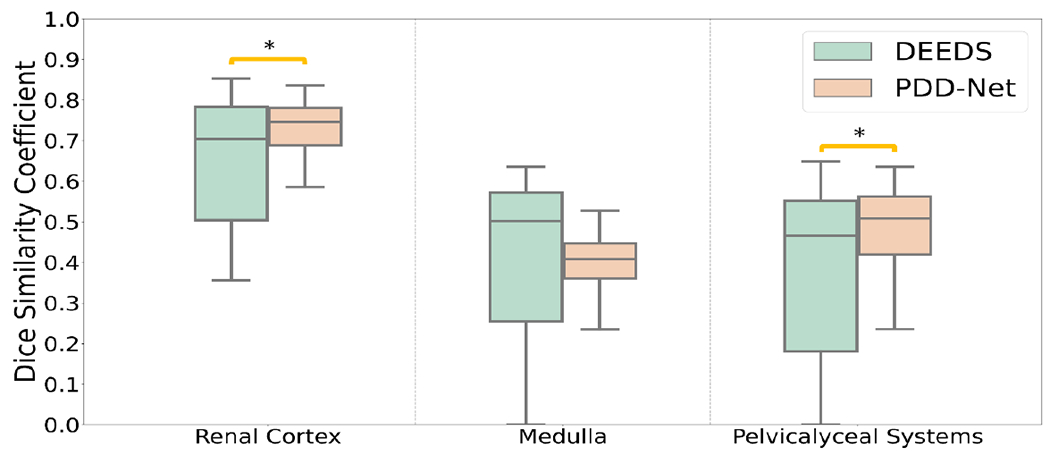
This quantitative representation demonstrates the registration performance from subject space to the atlas target with subject label transfer. Supervised registration performs with a substantial improvement in the renal cortex and pelvicalyceal system segmentation, while the performance of the medulla is limited with decrease of variance.
Apart from looking into the image registration performance, we evaluate the ability to adapt the significant morphology of kidney substructures by generating 2D color correspondence mapping with surface rendering. Here we compute 2D cielab checkerboard for each of the kidney substructure labels in the corresponding space and warp the checkerboard to the kidney substructure rendering, as shown in Figure 5. Each color represents the corresponding anatomical information located in the subject/atlas space. After warping with the displacement field, the checkerboard pattern follows the guidance of the displacement field and is deformed to demonstrate the adaptation of the subject information to the atlas space. The checkerboard pattern stably deformed from subject space to atlas space and well adapted the structural characteristics of the subject-wise kidney substructure. Grids in the pattern have not been overly deformed in the renal cortex labels, while a significant deformation is demonstrated to transfer the pelvicalyceal system information due to the significant variation of morphology.
Figure 5:
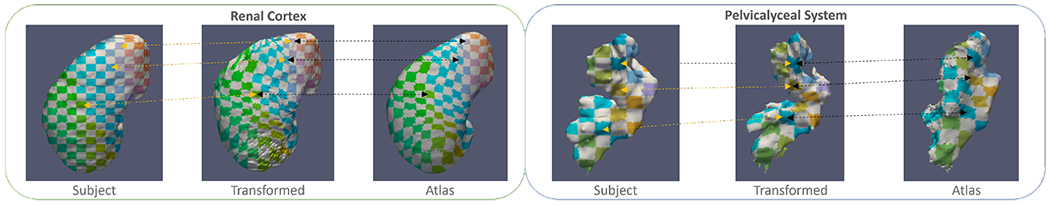
We illustrate the correspondence of the anatomical information in between the subjects’ space and atlas space after performing deep registration. Yellow arrows show the corresponding information of both organs before and after registration, while the black arrows correspond to the location of the registered context in both transformed images and the atlas target. The atlas template demonstrates stable adaptation of renal cortex context and significant deformation of pelvicalyceal system morphology across subject space.
5. DISCUSSION AND CONCLUSION
With the qualitative and quantitative representation above, the contrastive and morphological context of the kidney substructures is demonstrated with stability using the deep representation pipeline. The label transfer performance in the medullary regions of the kidney does not demonstrate significant improvement, although variance between registered labels is decreased in trend. As the deep registration model is trained with multi-organ labels, the registration aims to optimize the transformation of contextual information in the organ-corresponding regions, which is the complete kidney morphology, instead of the kidney substructures. With the opportunity of using label context in deep registration pipeline, we will adapt the current pre-train model to reduce the adverse effect from the morphological variability of other organs (such as the liver, spleen), and further fine-tune the model with the kidney substructure labels to optimize the kidney substructure registration. In this paper, we constructed a stable standard anatomical reference to localize the context of kidney substructure with deep network registration. The average mapping demonstrated the contrastive characteristics of each substructure across patients and the atlas target stably adapted the substructure information with the illustration of correspondence figure. We aim to create a minimal bias average template for healthy kidney substructures as our future long-term goal and analyze variability across populations.
6. ACKNOWLEDGEMENTS
This research is supported by NIH Common Fund and National Institute of Diabetes, Digestive and Kidney Diseases U54DK120058 (Spraggins), NSF CAREER 1452485, NIH 2R01EB006136, NIH 1R01EB017230 (Landman), and NIH R01NS09529. This study was in part using the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University, Nashville, TN. The identified datasets used for the analysis described were obtained from the Research Derivative (RD), database of clinical and related data. The imaging dataset(s) used for the analysis described were obtained from ImageVU, a research repository of medical imaging data and image-related metadata. ImageVU and RD are supported by the VICTR CTSA award (ULTR000445 from NCATS/NIH) and Vanderbilt University Medical Center institutional funding. ImageVU pilot work was also funded by PCORI (contract CDRN-1306-04869).
REFERENCES
- [1].Consortium H, “The human body at cellular resolution: the NIH Human Biomolecular Atlas Program,” Nature, vol. 574, no. 7777, p. 187, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rozenblatt-Rosen O, Stubbington MJ, Regev A, and Teichmann SA, “The Human Cell Atlas: from vision to reality,” Nature News, vol. 550, no. 7677, p. 451, 2017. [DOI] [PubMed] [Google Scholar]
- [3].Tang Y et al. , “Phase identification for dynamic CT enhancements with generative adversarial network,” Medical Physics, vol. 48, no. 3, pp. 1276–1285, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tang Y et al. , “Renal cortex, medulla and pelvicaliceal system segmentation on arterial phase CT images with random patch-based networks,” in Medical Imaging 2021: Image Processing, 2021, vol. 11596: International Society for Optics and Photonics, p. 115961D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gholipour A et al. , “A normative spatiotemporal MRI atlas of the fetal brain for automatic segmentation and analysis of early brain growth,” Scientific reports, vol. 7, no. 1, pp. 1–13, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Oishi K et al. , “Multi-contrast human neonatal brain atlas: application to normal neonate development analysis,” Neuroimage, vol. 56, no. 1, pp. 8–20, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].James GA, Hazaroglu O, and Bush KA, “A human brain atlas derived via n-cut parcellation of resting-state and task-based fMRI data,” Magnetic resonance imaging, vol. 34, no. 2, pp. 209–218, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rajashekar D et al. , “High-resolution T2-FLAIR and non-contrast CT brain atlas of the elderly,” Scientific Data, vol. 7, no. 1, pp. 1–7, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kuklisova-Murgasova M et al. , “A dynamic 4D probabilistic atlas of the developing brain,” NeuroImage, vol. 54, no. 4, pp. 2750–2763, 2011. [DOI] [PubMed] [Google Scholar]
- [10].Wang J, Zu H, Guo H, Bi R, Cheng Y, and Tamura S, “Patient-specific probabilistic atlas combining modified distance regularized level set for automatic liver segmentation in CT,” Computer Assisted Surgery, pp. 1–7, 2019. [DOI] [PubMed] [Google Scholar]
- [11].Li D et al. , “Augmenting atlas-based liver segmentation for radiotherapy treatment planning by incorporating image features proximal to the atlas contours,” Physics in Medicine & Biology, vol. 62, no. 1, p. 272, 2016. [DOI] [PubMed] [Google Scholar]
- [12].Heinrich MP, Maier O, and Handels H, “Multi-modal Multi-Atlas Segmentation using Discrete Optimisation and Self-Similarities,” VISCERAL Challenge@ ISBI, vol. 1390, p. 27, 2015. [Google Scholar]
- [13].Lee HH et al. , “Construction of a multi-phase contrast computed tomography kidney atlas,” in Medical Imaging 2021: Image Processing, 2021, vol. 11596: International Society for Optics and Photonics, p. 115961T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lee HH et al. , “Multi-Contrast Computed Tomography Healthy Kidney Atlas,” arXiv preprint arXiv:2012.12432, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Heinrich MP, Jenkinson M, Papież BW, Brady M, and Schnabel JA, “Towards realtime multimodal fusion for image-guided interventions using self-similarities,” in International conference on medical image computing and computer-assisted intervention, 2013: Springer, pp. 187–194. [DOI] [PubMed] [Google Scholar]
- [16].Heinrich MP, Jenkinson M, Brady M, and Schnabel JA, “MRF-based deformable registration and ventilation estimation of lung CT,” IEEE transactions on medical imaging, vol. 32, no. 7, pp. 1239–1248, 2013. [DOI] [PubMed] [Google Scholar]
- [17].Xu Z et al. , “Evaluation of six registration methods for the human abdomen on clinically acquired CT,” IEEE Transactions on Biomedical Engineering, vol. 63, no. 8, pp. 1563–1572, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tang Y et al. , “Body Part Regression With Self-Supervision,” IEEE Transactions on Medical Imaging, vol. 40, no. 5, pp. 1499–1507, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lee HH et al. , “Semi-supervised multi-organ segmentation through quality assurance supervision,” in Medical Imaging 2020: Image Processing, 2020, vol. 11313: International Society for Optics and Photonics, p. 113131I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Heinrich MP, “Closing the gap between deep and conventional image registration using probabilistic dense displacement networks,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2019: Springer, pp. 50–58. [Google Scholar]
- [21].Heinrich MP, Oktay O, and Bouteldja N, “OBELISK-Net: Fewer layers to solve 3D multi-organ segmentation with sparse deformable convolutions,” Medical image analysis, vol. 54, pp. 1–9, 2019. [DOI] [PubMed] [Google Scholar]
- [22].Dosovitskiy A et al. , “Flownet: Learning optical flow with convolutional networks,” in Proceedings of the IEEE international conference on computer vision, 2015, pp. 2758–2766. [Google Scholar]
- [23].Krähenbühl P and Koltun V, “Efficient inference in fully connected crfs with gaussian edge potentials,” Advances in neural information processing systems, vol. 24, pp. 109–117, 2011. [Google Scholar]
- [24].Felzenszwalb PF and Huttenlocher DP, “Efficient belief propagation for early vision,” International journal of computer vision, vol. 70, no. 1, pp. 41–54, 2006. [Google Scholar]
- [25].Jimenez-del-Toro O et al. , “Cloud-based evaluation of anatomical structure segmentation and landmark detection algorithms: VISCERAL anatomy benchmarks,” IEEE transactions on medical imaging, vol. 35, no. 11, pp. 2459–2475, 2016. [DOI] [PubMed] [Google Scholar]


