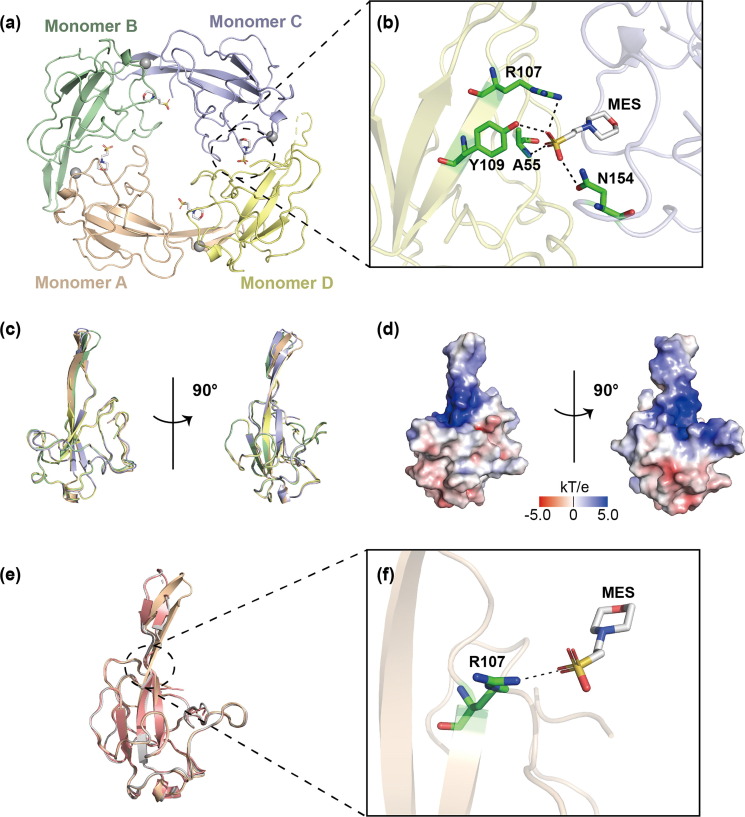
Fig. 1.
Crystal structure of SARS-CoV-2 N-NTD. (a) The overall structure of SARS-CoV-2 N-NTD tetramer (PDB ID: 7XX1). Each monomer is represented by a different color. (b) Hydrogen bonds between an MES molecule and different N-NTD monomers. Each monomer is represented by a different color. Hydrogen bonds are represented by black dotted lines, and amino acids involved in hydrogen bond interactions are marked. (c) The superimposition of four monomers in an asymmetric unit shown as a cartoon representation in two orientations. (d) The electrostatic potential surface of SARS-CoV-2 N-NTD in two orientations. Blue indicates positive charge potential, while red indicates negative charge potential. (e) Superposition (based on Cα) of our SARS-CoV-2 N-NTD structure (PDB ID: 7XX1, wheat) and other published SARS-CoV-2 N-NTD structures (PDB ID: 6M3M, gray; PDB ID: 7CDZ, pink). (f) Hydrogen bonds between an MES molecule and an N-NTD monomer. Hydrogen bonds are represented by black dotted lines, and amino acids involved in hydrogen bond interactions are marked.