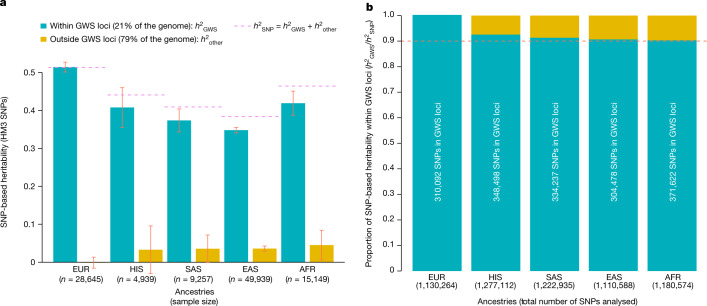
Fig. 3. Variance of height explained by HM3 SNPs within GWS loci.
a, Stratified SNP-based heritability () estimates obtained after partitioning the genome into SNPs within 35 kb of a GWS SNP ('GWS loci' label) versus SNPs that are more than 35 kb away from any GWS SNP. Analyses were performed in samples of five different ancestries or ethnic groups: European (EUR: meta-analysis of UK Biobank (UKB) + Lifelines study), African (AFR: meta-analysis of UKB + PAGE study), East Asian (EAS: meta-analysis of UKB + China Kadoorie Biobank), South Asian (SAS: UKB) and Hispanic (HIS: PAGE). Error bars represent standard errors. b, More than 90% of in all ancestries is explained by SNPs within GWS loci identified in this study. The cumulative length of non-overlapping GWS loci is around 647 Mb; that is, around 21% of the genome, assuming a genome length of around 3,039 Mb (ref. 26). The proportion of HM3 SNPs in GWS loci is around 27%.