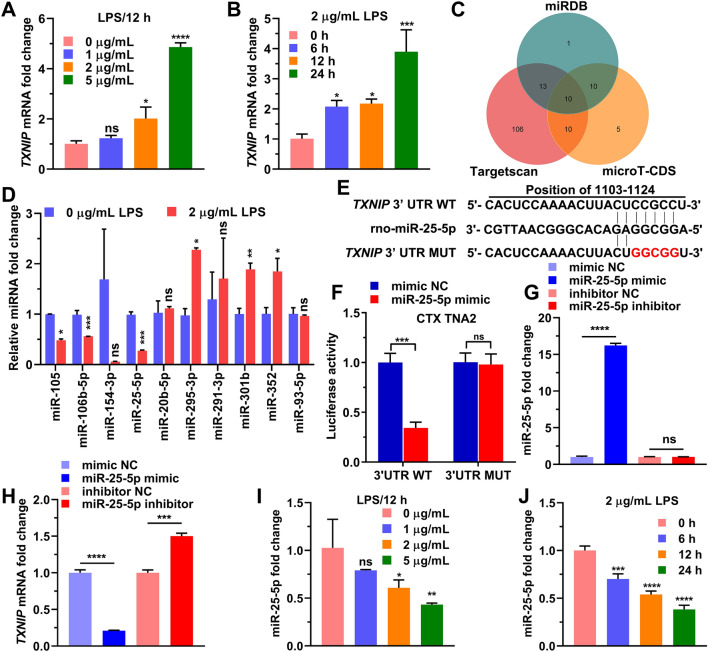
Figure 1.
miR-25-5p negatively regulates TXNIP expression. (A) TXNIP fold change of CTX TNA2 cells in response to 0 μg/mL, 1 μg/mL, 2 μg/mL, and 5 μg/mL LPS stimulation over 12 h. (B) TXNIP fold change of CTX TNA2 cells in response to 2 μg/mL LPS stimulation over 0 h, 6 h, 12 h, and 24 h. (C) Numbers of miRNAs predicted to bind to 3′-UTR sequence of TXNIP mRNA from miRDB, microT-CDS, and Targetscan. (D) The fold changes of top 10 predicted miRNAs CTX TNA2 cells in response to 2 μg/mL LPS stimulation. (E) Sequences of wild-type (TXNIP 3′UTR WT) and mutant TXNIP 3′-UTR (TXNIP 3′UTR MUT). (F) Luciferase activities of recombined reporter plasmids (TXNIP 3′UTR WT and TXNIP 3′UTR MUT) co-transfected with miR-25-5p mimic. (G, H) miR-25-5p (G) and TXNIP (H) mRNA fold change of CTX TNA2 cells in response to the transfection of miR-25-5p mimic and miR-25-5p inhibitor. (I) miR-25-5p fold change of CTX TNA2 cells in response to 0 μg/mL, 1 μg/mL, 2 μg/mL, and 5 μg/mL LPS stimulation over 12 h. (J) miR-25-5p fold change of CTX TNA2 cells in response to 2 μg/mL LPS stimulation over 0 h, 6 h, 12 h, and 24 h. The relative expression levels of miRNA and mRNA were determined by qPCR and protein levels were determined by western blotting. The independent t-test was applied to the comparison between groups (D, F, G, and H). One-way analysis of variance (ANOVA), followed by Tukey’s test was used to identify the significant differences among multiple groups (A, B, I, and J). Values are shown as the mean ± standard deviation (SD). Con: negative control of LPS, LPS: lipopolysaccharide, mimic NC: negative control of miR-25-5p mimic, inhibitor NC: negative control of miR-25-5p inhibitor. ns indicates not significant. *P < 0.05. **P < 0.01. ***P < 0.001. ****P < 0.0001.