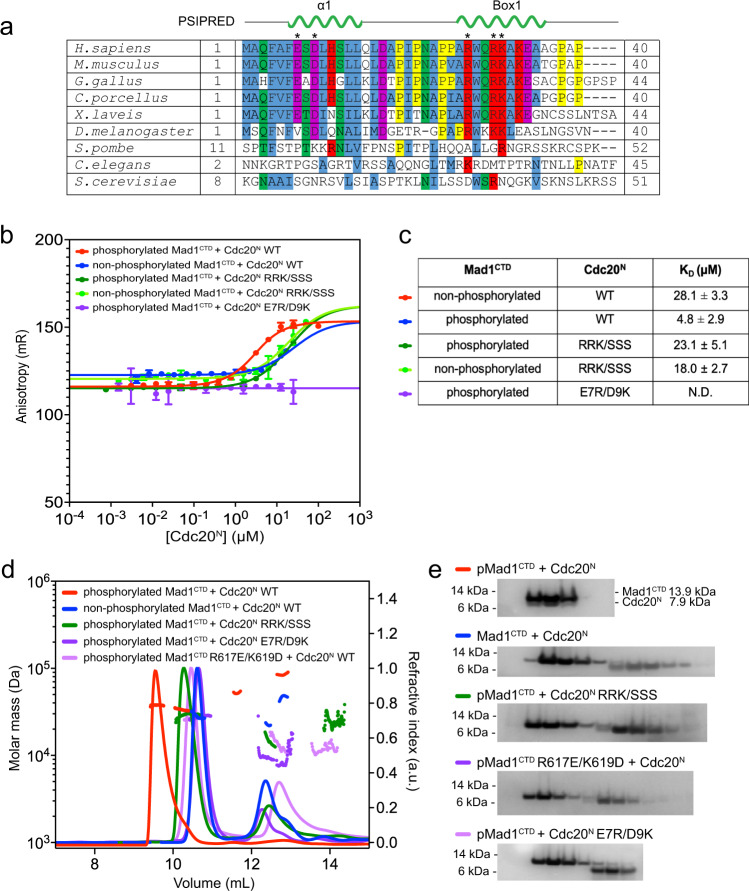
Fig. 5. Binding of Cdc20N and Mad1CTD mutants by fluorescent anisotropy and SEC-MALS.
a A multiple sequence alignment of Cdc201–40. Secondary structure prediction by PSIPRED is shown above for the α1 and Box1 predicted helices. The asterisks denote residues which were mutated and tested by fluorescent anisotropy or SEC-MALS in b–d. b Fluorescent anisotropy with preformed AF488-Bub1CD1:Mad1CTD complex (20 nM: 2.1 µM) using pMad1CTD or Mad1CTD and where various Cdc20N mutants were titrated. The data were presented as mean values ± SD derived from three independent measurements. c Summary of the calculated KD of the interactions analysed in (b). d SEC-MALS analysis of Cdc20N interaction with pMad1CTD or Mad1CTD using various mutants. Theoretical masses for Mad1CTD dimer, Cdc20N, and Mad1CTD dimer with a single Cdc20N bound are 28, 7.8 and 36 kDa respectively. e SDS-PAGE analysis of the SEC-MALS experiments shown in (d), using a 4–12% Bis-Tris Glycine gel with the SeeBlue™ Plus2 Protein Ladder (Thermo Fisher Scientific). Three independent measurements of each sample were completed with similar results.