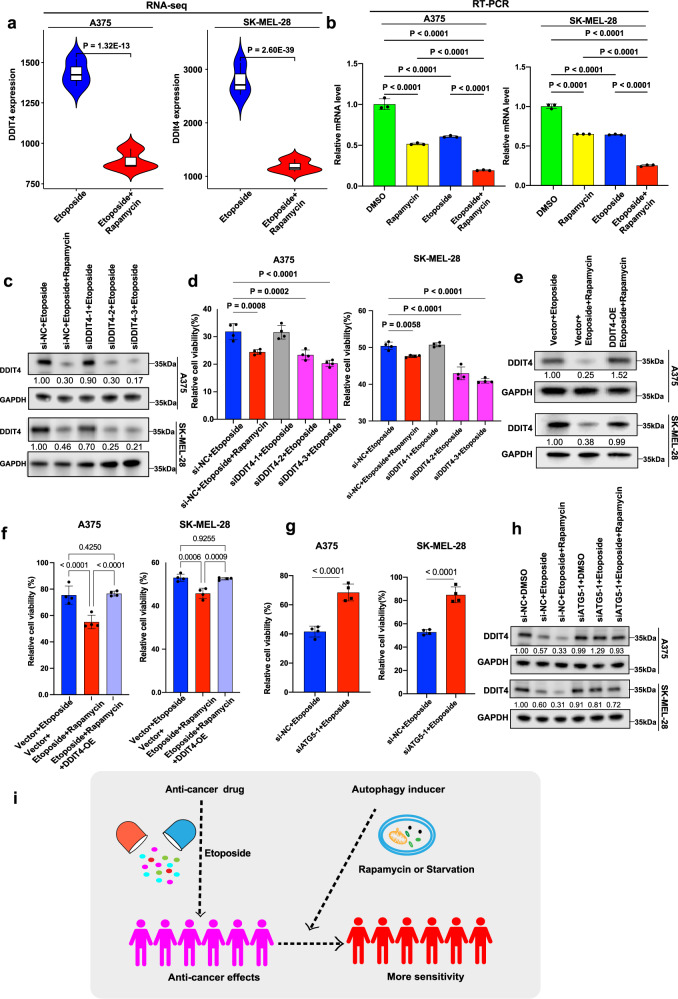
Fig. 8. Potential mechanism through the functional characterization of DDIT4 in vitro.
a The mRNA expression of DDIT4 in A375 and SK-MEL-28 by RNA-seq (n = 3). The boxes show the median ±1 quartile, with whiskers extending to the most extreme data point within 1.5 interquartile range from the box boundaries. The significance (p value) of differentially expressed genes was evaluated with ‘DESeq2’ package. b Relative mRNA expression of DDIT4 in A375 and SK-MEL-28 by RT-PCR (n = 3). c Western blot of DDIT4 in A375 or SK-MEL-28 cells treated with si-NC or DDIT4 siRNAs in combination with etoposide. DDIT4 band intensities normalized to si-NC + etoposide are displayed below the blots. d The cell viability of A375 or SK-MEL-28 cells treated with si-NC or DDIT4 siRNAs in combination with etoposide (n = 4). Pink bars denote the successful knockdown of si-DDIT4 (si-DDIT4-2 and si-DDIT4-3), while gray bar denotes the unsuccessful knockdown of si-DDIT4 (si-DDIT4-1). e Western blot of DDIT4 in A375 or SK-MEL-28 cells transfected with DDIT4-OE lentivirus or vector in combination with etoposide and rapamycin. DDIT4 band intensities normalized to Vector+Etoposide are displayed below the blots. f The cell viability of A375 or SK-MEL-28 cells transfected with DDIT4-OE lentivirus or vector in combination with etoposide and rapamycin (n = 4). g The cell viability of A375 or SK-MEL-28 cells treated with si-NC or ATG5 siRNA in combination with etoposide (n = 4). h Western blot of DDIT4 in A375 or SK-MEL-28 cells treated with si-NC or ATG5 siRNAs in combination with etoposide. DDIT4 band intensities normalized to si-NC + DMSO are displayed below the blots. i The illustration for the potential mechanism of autophagy inducer sensitizing drug response. b, d, f, g Data were presented as means ± SD. b, d, f The difference in multiple groups was estimated by one-way ANOVA analysis. g Two-sided student’s t test was used for the estimation of the difference in two groups. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns is not significant. c, e, h See Supplementary Fig. 8 for uncropped data.