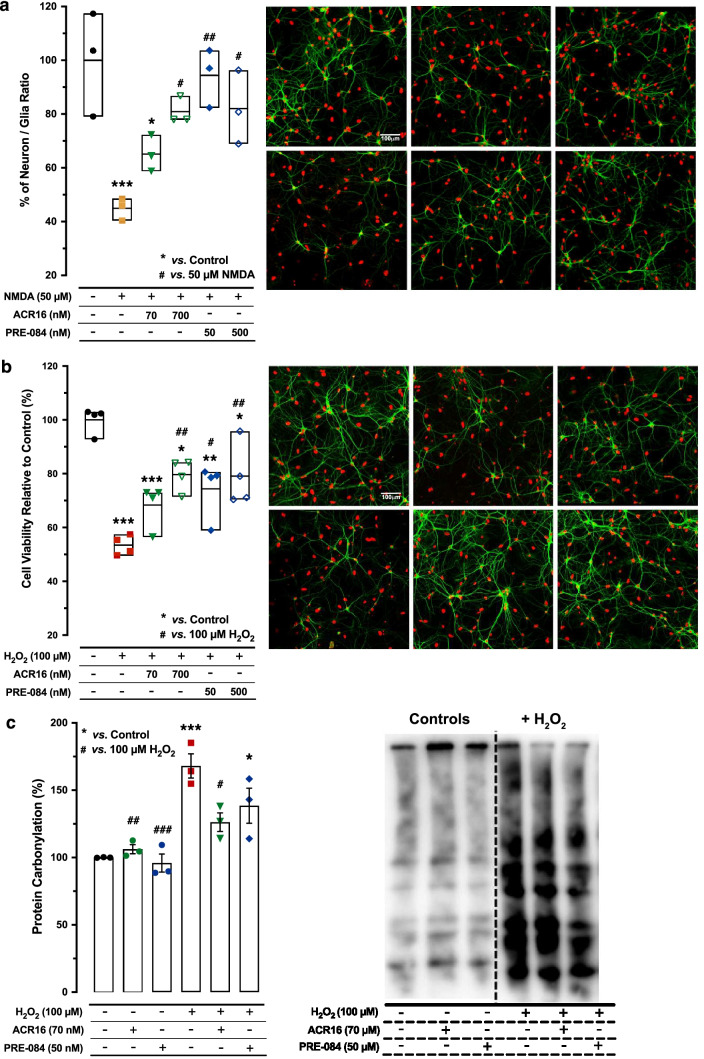
Fig. 1.
ACR16 and PRE-084 protect against cell death from oxidative stress and NMDA-related toxicity in primary hippocampal cultures. a Left: quantitative summary of neuronal survival in NMDA-induced toxicity model and following 24 h of exposure (n = 3 for each), as indicated. Data are expressed as a percentage over control conditions [expressed as mean (Min–Max)]. Analysis by 1-way ANOVA (F (5,12) = 9.485; ***p < 0.001) and Sidak’s multiple comparisons test. Right: confocal fluorescence microscopy images from rat hippocampal primary cell cultures in which neurons were identified and labeled by MAP-2 immunocytochemistry (green) and cell nuclei were stained with DAPI (pseudocolored in red). Scale bar = 100 μm. b Left: quantitative summary that expresses cell viability as the percentage of cell densities relative to those in control conditions following 24 h of exposure [mean (Min–Max); n = 4 for each), as indicated. Data were analyzed using 1w-ANOVA (F(5,18) = 14.75; ***p < 0.001) and Sidak’s multiple comparisons test. Right: confocal fluorescence microscopy images from rat hippocampal primary cell cultures following 24 h of exposure to different treatments in the H2O2-induced toxicity model, as indicated. Neurons were identified by MAP-2 immunocytochemistry (green), while cell nuclei were stained using DAPI (red). Scale bar = 100 μm. Significant differences compared with respect to untreated controls are displayed as *, **, and *** for p < 0.05, p < 0.01, and p < 0.001, respectively. Similarly, comparisons again toxicity treatments alone, NMDA (a) or H2O2 (b), are shown as #, ##, and ### indicating significant differences of p < 0.05, p < 0.01, and p < 0.001, respectively. Note that the difference between * and # was also indicated directly in the panel. c Left: quantitative summary of carbonylated status of proteins in 12 DIV rat hippocampal primary cell cultures following exposure of the indicated treatments for 24 h (mean ± SEM; n = 3 for each). Right: representative immunoblot using the Oxyblot kit for detection of carbonylated proteins. Analysis by 2-way ANOVA (F (2,12) = 4.889; *p < 0.05) followed by Tukey’s multiple comparisons test was used here for data analysis. Difference between * and # was also indicated directly in this panel