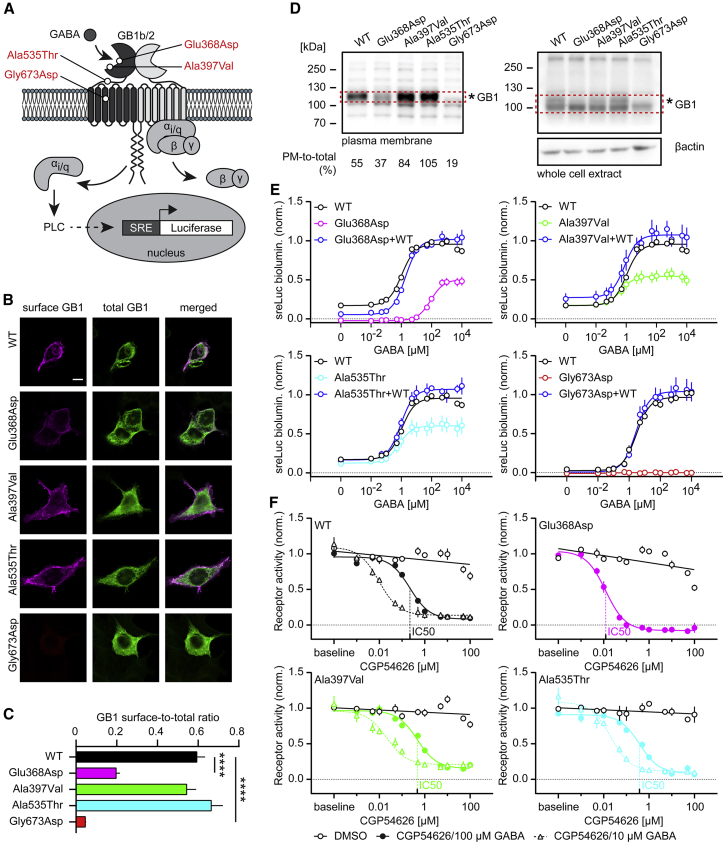
Figure 1.
Functional characterization of de novo GB1 variants in HEK293 cells
(A) Location of GB1 variants in the heterodimeric GBR. The scheme additionally depicts the assay used to monitor PLC-dependent RLuc expression under control of the serum response element (SRE). GB1b/2 receptors were artificially coupled to PLC by stable expression of the chimeric G protein subunit Gαqi in HEK293T cells. GB1b, GB2, and SRE-RLuc were transiently transfected.
(B) Surface and total GB1b expression levels of WT and mutant HA-tagged GB1b subunits co-expressed with GB2 in transfected HEK293 cells. Surface GB1b expression levels were determined with anti-HA antibodies in non-permeabilized cells. Total GB1b expression levels were determined with anti-GB1 antibodies in permeabilized cells. Scale bar: 10 μm.
(C) Bar graphs of the GB1b surface-to-total expression ratio. Kruskal-Wallis (p < 0.0001) and Dunn’s multiple comparison tests were used because of failed Shapiro-Wilk normality test. ∗∗∗∗p < 0.0001; WT, n = 28; p.Ala397Val, n = 38; p.Ala535Thr, n = 32; p.Glu368Asp, n = 32; p.Gly673Asp, n = 26.
(D) Fractionation of plasma membrane (PM) proteins in HEK293 cells transiently expressing GB1b/2 receptors. GB1 proteins were identified on immunoblots. WT and variant GB1 protein in the red rectangles was quantified densitometrically. The percentage of PM-to-total GB1 protein is indicated. Immunostaining for β-actin was used to control for loading. The results are from a single experiment. (∗) indicates the glycosylated form of GB1. Of note, the loss of glycosylated plasma membrane GB1 protein observed with the p.Glu368Asp and p.Gly673Asp variants is consistent with impaired maturation and surface transport of these proteins.
(E) Dose-response curves showing GABA-induced RLuc activity by WT, variant, and WT+variant (1:1 GB1b plasmid DNA ratio) GBRs in transfected cells. Three parameter log(GABA) versus response nonlinear regression curve fit was used for GABA dose response curves, with the exception of p.Gly673Asp, for which a linear regression curve fit was used. Table S1 provides information about EC50, Emax, and basal activity values derived from the GABA concentration-response curves and a statistical analysis.
(F) Dose-response curves showing inhibition of GBR activity (10 μM or 100 μM GABA) by the antagonist CGP54626. Three parameter log(inhibitor) versus response nonlinear regression curve fit was used for CGP54626 dose-response curves and linear regression curve fits for the DMSO controls. The curve fits depict DMSO controls for 100 μM GABA only. All data are mean ± SEM. The number of independent experiments is indicated. Table S2 provides IC50 values and a statistical analysis of the CGP54626 inhibition curves.