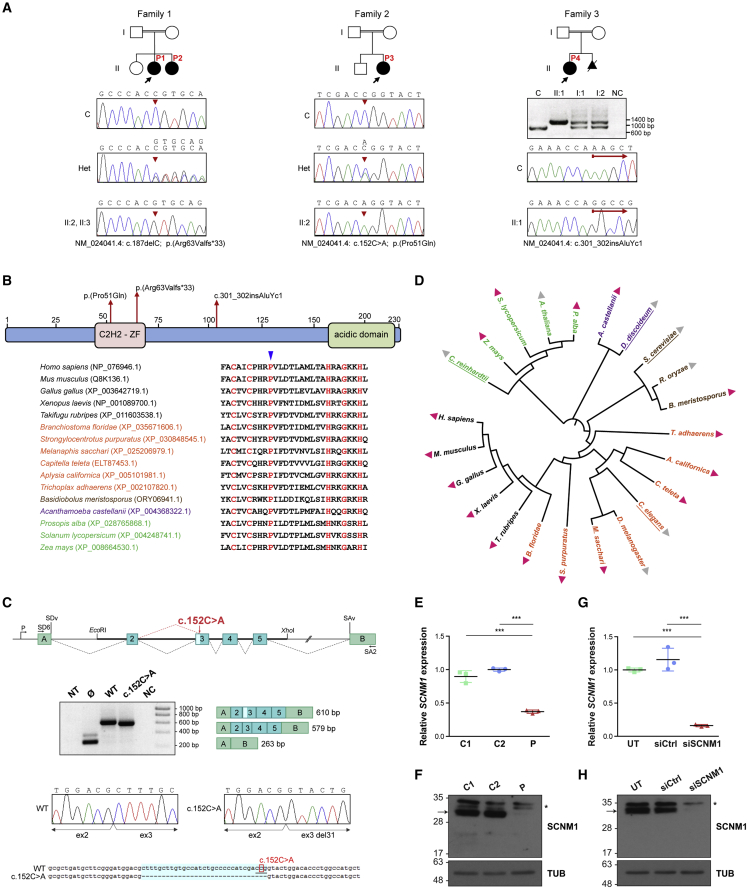
Figure 2.
Identification of loss-of-function mutations in SCNM1 in individuals with OFD
(A) Family pedigrees of affected individuals P1–P4 described in this report. Probands are designated with black arrows. Genomic DNA sequence chromatograms of SCNM1 illustrating the homozygous mutation (red arrowheads) identified in each family are displayed underneath. The sequence of affected individuals, heterozygous carriers (Het), and a control sample (C) are shown. Nucleotide sequences corresponding to mutant (top) and normal (underneath) alleles are written on Het electropherograms. The agarose gel image below the pedigree of family 3 shows the PCR products resulting from the amplification of a genomic DNA fragment containing SCNM1 exon 4 in P4 (II:1), both of her parents (I:1, I:2), and in a control individual (C). NC, no DNA control. The red arrows in family 3 chromatograms designate the site of the AluYc1 insertion after which control and P4 DNA sequences diverge.
(B) Schematic representation of the SCNM1 protein indicating the position of the C2H2 zinc finger (C2H2-ZF) domain and the acidic domain of this protein, and the location of the three mutations indicated in (A). A sequence alignment of the C2H2-ZF domain from different SCNM1 orthologs showing high degree of conservation of Pro51 (blue arrowhead), is also indicated.
(C) SCNM1 minigene assay. Top: schematic representation of pSPL3/SCNM1 hybrid minigene. Boxes are exons and dotted lines connecting exons indicate normal (black) or altered (red) splicing events. The two artificial exons (A and B), promoter (P), donor (SDv), and acceptor (SAv) splice sites of pSPL3 are depicted. EcoRI and XhoI restriction sites used to clone the wild-type (WT) and c.152C>A SCNM1 genomic fragments as well as primers SD6 and SA2 used for RT-PCR are shown. Middle: representative gel electrophoresis image (n = 3) showing RT-PCR products obtained in cells transfected with the WT or the c.152C>A minigene or the empty vector (Ø). NT, non-transfected; NC, no cDNA control. Exon composition and sizes (WT [610 bp] and c.152C>A [579 bp]) of the amplified products are on the right. The light blue box in exon 3 of the WT PCR product represents the 31 bp that are missing in the c.152C>A PCR fragment. Sanger sequencing chromatograms of WT and c.152C>A RT-PCR products show the loss of the first 31 bp of SCNM1 exon 3 in the product from the mutant minigene. Bottom: DNA sequence alignment of WT and c.152C>A RT-PCR products highlighting (light blue) the 31 nucleotides missing in the c.152C>A sequence. Note the new CAG acceptor splice site (underlined nucleotides) created by the c.152C>A mutation (boxed nucleotide).
(D) Common taxonomy tree of representative eukaryotic species from distantly related phylogenetic taxa. Presence and absence of SCNM1 is indicated with pink and gray triangles, respectively. Species previously reported with no minor introns are underlined. Protists, fungi, algae and plants, and invertebrate and vertebrate animals are indicated in different colors. Full names and taxa of species are listed in material and methods.
(E) Relative quantification of SCNM1 expression by RT-qPCR in fibroblasts from controls (C1, C2) and affected individual P2 (P). Gene expression was normalized against the geometric mean of GAPDH and GUSB mRNA levels, and the ΔCt mean value of C2 was used as calibrator sample. Data are expressed as mean ± SD (n = 3). ∗∗∗p < 0.001. One-way ANOVA with Tukey’s multiple comparison test.
(F) Representative anti-SCNM1 immunoblot (n = 3) showing absence of SCNM1 in fibroblasts from P2 (P). Control fibroblasts: C1, C2. SCNM1 is indicated with an arrow, and the asterisk designates nonspecific bands. Tubulin (TUB) was used as a loading control.
(G) Relative quantification of SCNM1 expression (n = 3) by RT-qPCR in RPE-1 cells untransfected (UT) or transfected with non-targeting siRNA (siCtrl) or with siRNA against SCNM1 (siSCNM1). Gene expression analysis was performed as in (E) using the ΔCt mean value of UT cells as the calibrator sample. Data are expressed as mean ± SD (n = 3). ∗∗∗p < 0.001. One-way ANOVA with Tukey’s multiple comparison test.
(H) Representative anti-SCNM1 immunoblotting (n = 4) showing reduced levels of the SCNM1 protein in SCNM1-KD RPE-1 cells (siSCNM1) compared to UT and siCtrl cells. SCNM1 is indicated with an arrow, and the asterisk designates a nonspecific band. Loading control: tubulin (TUB).