Figure 4.
Biochemical characterization of the TMEM147 mutant proteins and immunostainings in fibroblasts
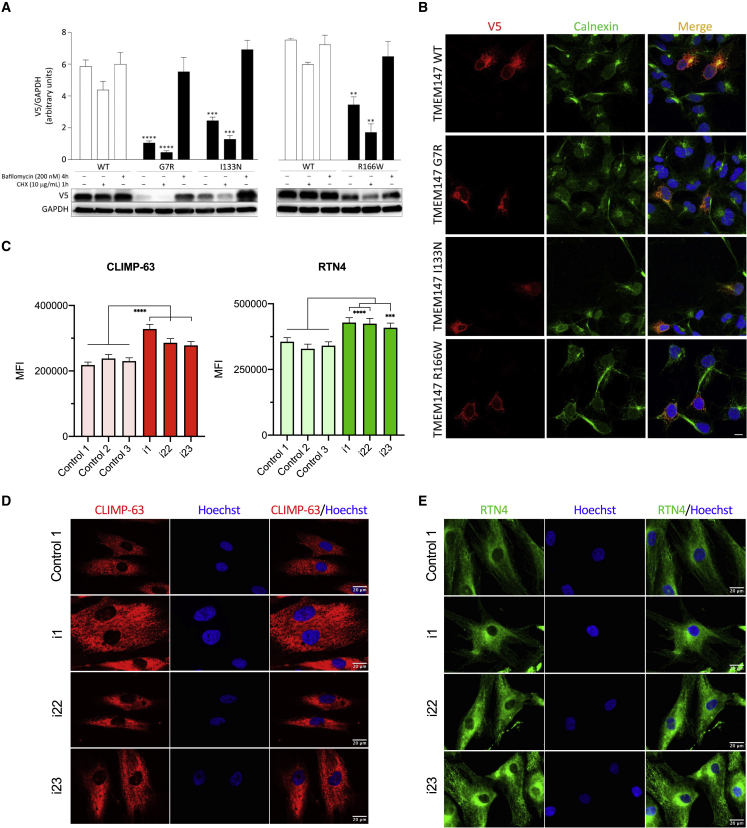
(A) Accelerated degradation of the TMEM147Arg7Gly (R7G), TMEM147Ile133Asn (I133N), and TMEM147Arg166Trp (R166W) proteins. Immunoblot analysis shows WT and variant V5-tagged TMEM147 protein levels in transfected COS-1 cells, basally and after CHX (10 μg/mL) or bafilomycin (200 nM) treatment. GAPDH was used as loading control. Representative blots (below) and mean ± SD densitometry values (above) of three independent experiments are shown. Asterisks indicate statistically significant differences compared with WT TMEM147 (∗∗∗∗p ≤ 0.0001; ∗∗∗p ≤ 0.001; ∗∗p ≤ 0.05; two-way ANOVA followed by Tukey’s multiple comparison test).
(B) Subcellular localization of transiently expressed V5-tagged WT or mutant TMEM147 proteins in COS-1 cells under steady-state conditions revealed by confocal microscopy analysis. Cells were stained with the anti-V5 monoclonal antibody (red). Co-localization analysis was performed using the endoplasmic reticulum marker calnexin (green). Merged images with nuclei (Hoechst 33342 staining, blue) are displayed on the right. Scale bar, 10 μm.
(C) Quantification of mean fluorescence signals ± SEM detected in (D). Three technical replicates were performed per cell line. A total of 150 measurements per cell line were performed. Asterisks indicate statistically significant differences compared with cell lines from healthy individuals: control 1, control 2, and control 3 (∗∗∗∗p ≤ 0.0001; ∗∗∗p ≤ 0.001; ∗∗p ≤ 0.01; ∗p ≤ 0.05; one-sample Wilcoxon test, based on the average of the three control samples).
(D and E) Localization analysis of CKAP4 (CLIMP-63) (D) and RTN4 (E) in fibroblasts from i1, i22, and i23 and healthy control individuals (only control 1 is shown in the figure). Scale bar, 20 μm.