Figure 5.
RGD neurons displayed aberrant electrophysiological properties
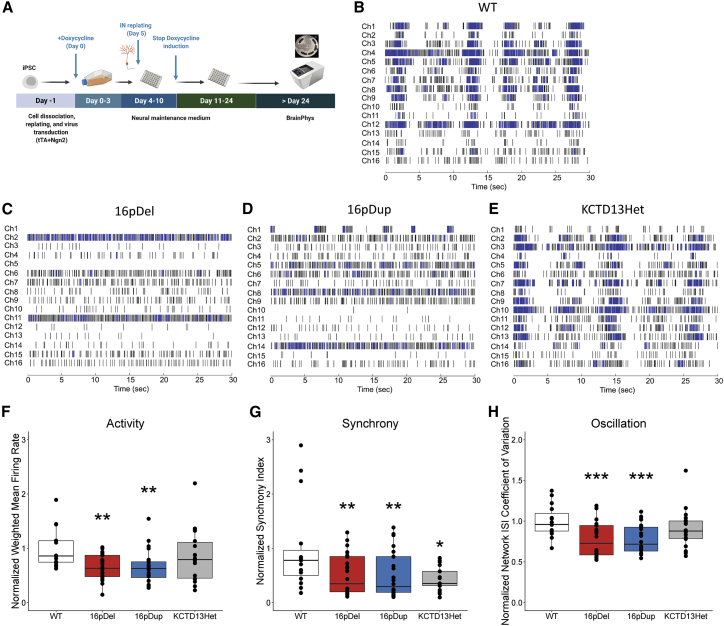
(A) Overview of the study design. iNs were differentiated from hiPSCs as described in the methods section. On day, 5 iNs were replated onto MEA plates with NMM. The neural activities were recorded after the culture medium switched to BrainPhys Neuronal Medium (day 24).
(B–E) Representative temporal raster plots from iN models demonstrating the activity over time for all electrodes in the well. Each plot is 30 s for sufficient spike and burst resolution, and horizontal rows correspond to 16 electrodes. Abbreviations are as follows: wild type (WT), 16p11.2 deletion (16pDel), 16p11.2 duplication (16pDup), and KCTD13 heterozygous deletion (KCTD13Het). Raster plots were generated with Neural Metric Tool v3.2.5 software (Axion Biosystems).
(F) Neuron activity (normalized weighted-mean firing rate). 16pDel and 16pDup neurons displayed significantly lower activity than WT neurons (∗∗p < 0.01), whereas KCTD13Het neurons displayed a level of activity comparable to that of the WT (p = 0.222). Data are presented as means ± SEM, and normalized data points are plotted.
(G) Neuron synchrony (normalized synchrony Index). 16pDel, 16pDup, and KCTD13Het neurons displayed significantly lower synchrony than WT neurons (∗p < 0.05, ∗∗p < 0.01). Data are presented as means ± SEM, and normalized data points are plotted.
(H) Neuron network oscillation (normalized network ISI coefficient of variation). 16pDel and 16pDup neurons displayed significantly lower oscillation than WT neurons (∗∗∗p < 0.001), whereas KCTD13Het neurons displayed a level of activity comparable to that of the WT (p = 0.276). The number of samples per group was n = 15 (WT), n = 24 (16pDel), n = 24 (16pDup), and n = 18 (KCTD13Het). Data are presented as means ± SEM, and normalized data points are plotted.