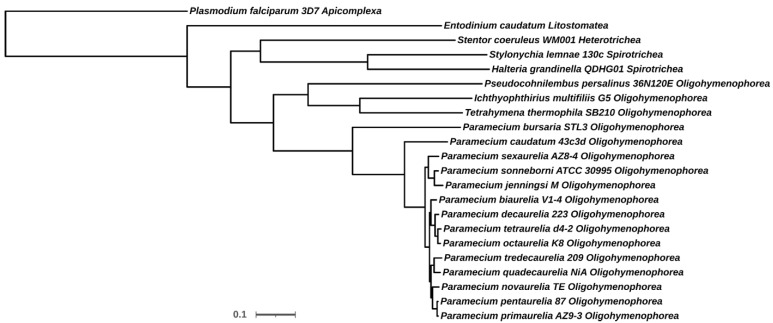
Figure 1.
Phylogenetic tree of selected species of ciliates. Protein datasets were downloaded either from ParameciumDB or NCBI, except for Entodinium caudatum, which was kindly provided by Drs. Yu and Park from Ohio State University, OH, USA. All proteomes were used for feeding Orthofinder (see text above for reference). A total of 507 orthogroups with all the species was used to build the tree, using the STAG algorithm included in Orthofinder. The inferred tree was drawn with iTOOL [36]. P. falciparum, an apicomplexan close relative of ciliates, was selected as outgroup by the STRIDE algorithm, also included in Orthofinder. For each species, the full name, strain, and Class to which each species belongs are shown, except for P. falciparum, for which the Phylum was written instead of the Class. The scale bar represents the average number of substitutions per sites across of the 507 orthogroups used.