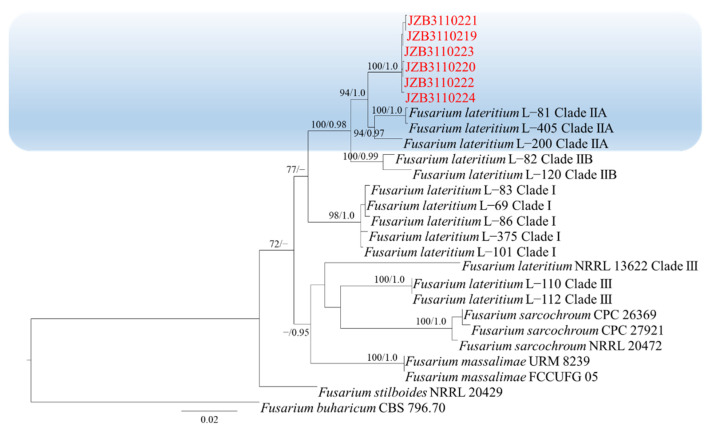
Figure 3.
Phylogenetic tree generated by maximum likelihood analysis (RAxML) of FLSC based on the combined rpb2, tef1 and tub2 sequence data. The tree is rooted with Fusarium buharicum (CBS 796.70). The best-scoring RAxML tree with a final likelihood value of −5602.556442 is presented. The matrix had 358 distinct alignment patterns, with 32.15% being undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.236754, C = 0.282131, G = 0.242563, T = 0.238552; substitution rates AC = 2.260928, AG = 7.239234, AT = 2.341489, CG = 1.331525, CT = 16.787035, GT = 1.000000; gamma distribution shape parameter α = 1.252177. ML bootstrap support values ≥50% and Bayesian posterior probabilities (BYPP) ≥ 0.95 are given near the nodes. The scale bar indicates 0.02 changes per site. Isolates belonging to this study are given in red.