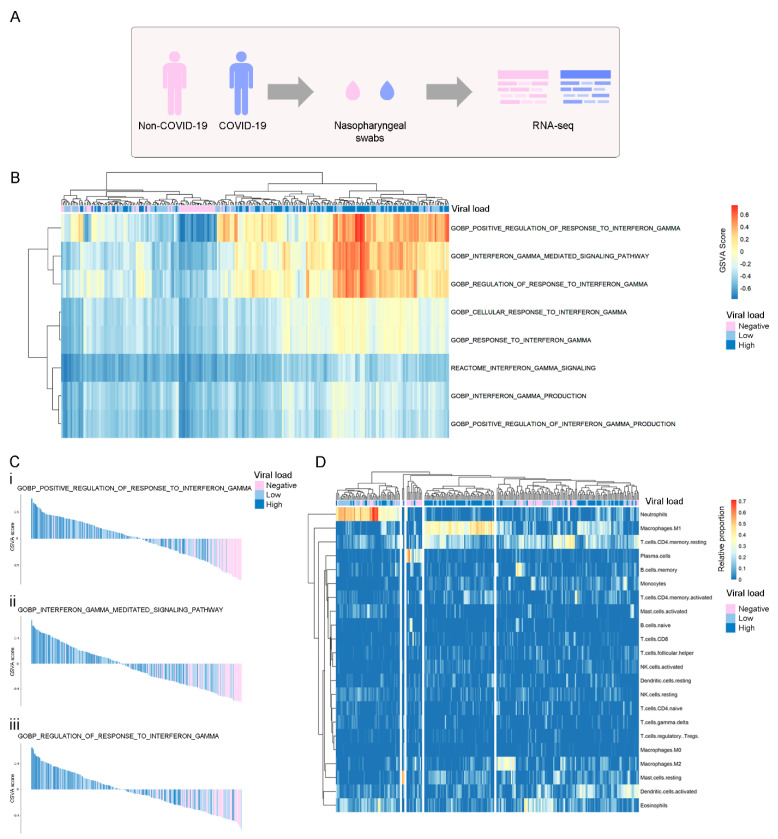
Figure 1.
Global assessment at the transcriptional level of pathways and immune cell types related to IFN-γ production, signaling, and regulation of response in COVID-19-positive (light blue and blue) and -negative (pink) patients. (A) Experimental design of the GSE152075 dataset, composed of transcriptome data from nasopharyngeal swabs collected from 430 COVID-19 and 54 non-COVID-19 patients. (B) Non-supervised clustering of patients according to their GSVA score in each IFN-γ geneset. Higher GSVA scores indicate higher activity of the geneset at the RNA level. Each column is labeled according to the COVID-19 viral load of each patient (negative: pink, low: light blue, high: blue). (C) Waterfall plots of selected genesets that were highly activated in COVID-19 patients vs. non-COVID-19 patients. Patients are ordered from the highest to the lowest GSVA score in each geneset. (D) Unsupervised clustering of non-COVID-19 and COVID-19 patients according to the relative proportions of immune cell types estimated by CIBERSORT (LM22 signature) in RNA-seq data. Each column is labeled according to the COVID-19 viral load of each patient. Viral load is represented as a color scale and was categorized as Negative (pink), Low (first quartile; light blue), or High (fourth quartile; blue). COVID-19 patients with intermediate viral load were excluded from the analysis.