FIGURE 1.
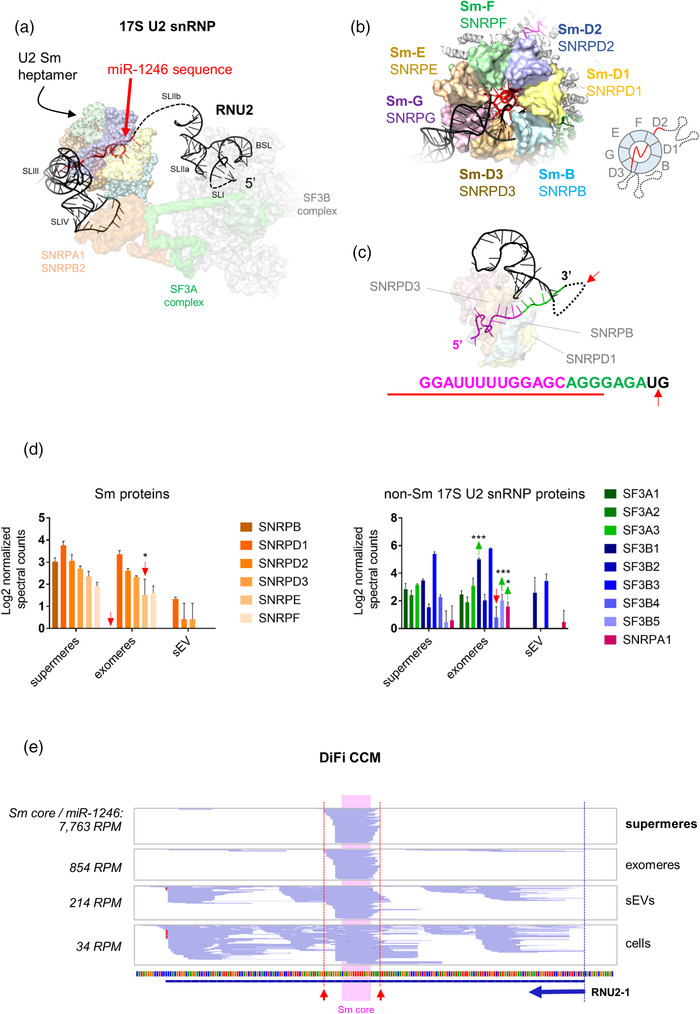
Extracellular miR‐1246 corresponds to the footprint of the Sm heptamer protecting the oligo(U) sequence motif of the U2 small nuclear RNA. (a) Cryo‐EM structure of the 17S U2 snRNP (PDB: 6Y5Q) showing the sequence of miR‐1246 in red. (b) Three‐dimensional model of the Sm heptamer, forming a ring around the miR‐1246‐like sequence of RNU2 (based on the structure of the precursor pre‐catalytic spliceosome, PDB: 6AH0). (c) The sequence of RNU2 protected by the Sm ring is shown in magenta. Purines extending toward the 3′ end are shown in green. The red arrow represents the putative cleavage site by members of the RNase A superfamily. The red line represents the sequence of the mature hsa‐miR‐1246 according to miRBase. (d) Normalised spectral counts of Sm proteins (left) and U2‐associated splicing factors (right) in supermeres, exomeres and EVs from DiFi cells, based on data provided in Zhang et al. (2021). Proteins showing statistically higher (green arrows) or lower (red arrows) normalised spectral counts in exomeres versus supermeres are indicated (two‐way ANOVA with Tukey's multiple comparison test). * p < 0.05. *** p < 0.001. (e) Coverage plots of RNU2‐1 in supermeres, exomeres, EVs and DiFi cells (replicate 1), based on small RNA sequencing data provided in Zhang et al. (2021). The blue arrow indicates the start and direction of RNU2‐1. Note the blue‐coloured reads correspond to the minus DNA strand, and their orientation (5′‐3′) is therefore right‐to‐left. Red arrows indicate predictive cleavage sites (by RNase A family members) 3′ to the most proximal pyrimidines outside of the Sm core region.
