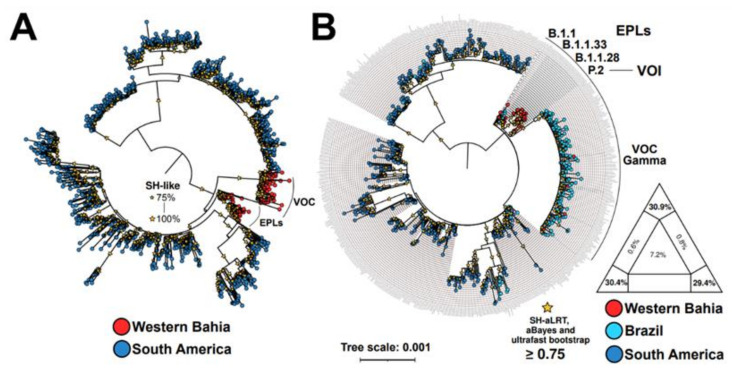
Figure 5.
Maximum-likelihood midpoint rooted phylogenetic tree based on 1117 (A) and 603 (B) representative genome sequences of SARS-CoV-2. Nextstrain’s subsampled SARS-CoV-2 genomic data from South America (including Brazil) since pandemic started up to August 2022 containing unfiltered (A) and filtered (B) sequences from the study. The SARS-CoV-2 genomes from this study are identified by the red circles. Tips are colored according to sampling locations. Yellow stars assume Shimodaira–Hasegawa (SH-like) test (A) and SH-aLTR/aBayes/ultrafast bootstrap support (B) based on 1000 replicates. Only values equal or greater than 75% are shown. Likelihood mapping of the final sequences alignment showing low phylogenetic noise, as required for reliable phylogeny inference (B). Abbreviations: VOC and VOI, Variant(s) of Concern and Variant(s) of Interest, respectively. EPLs, Early Pandemic Lineages. In letter B, “Brazil” represents whole-genome of SARS-CoV-2 Gamma variant from all Brazilian States and the Federal District. Branch lengths are drawn in scale of nucleotide substitutions per site according to the bar scale. Colors and symbols used in the panels are defined according to the legend to the left and right of the figure.