Figure 2: Repeat-induced transcriptional silencing, R-loops and somatic instability.
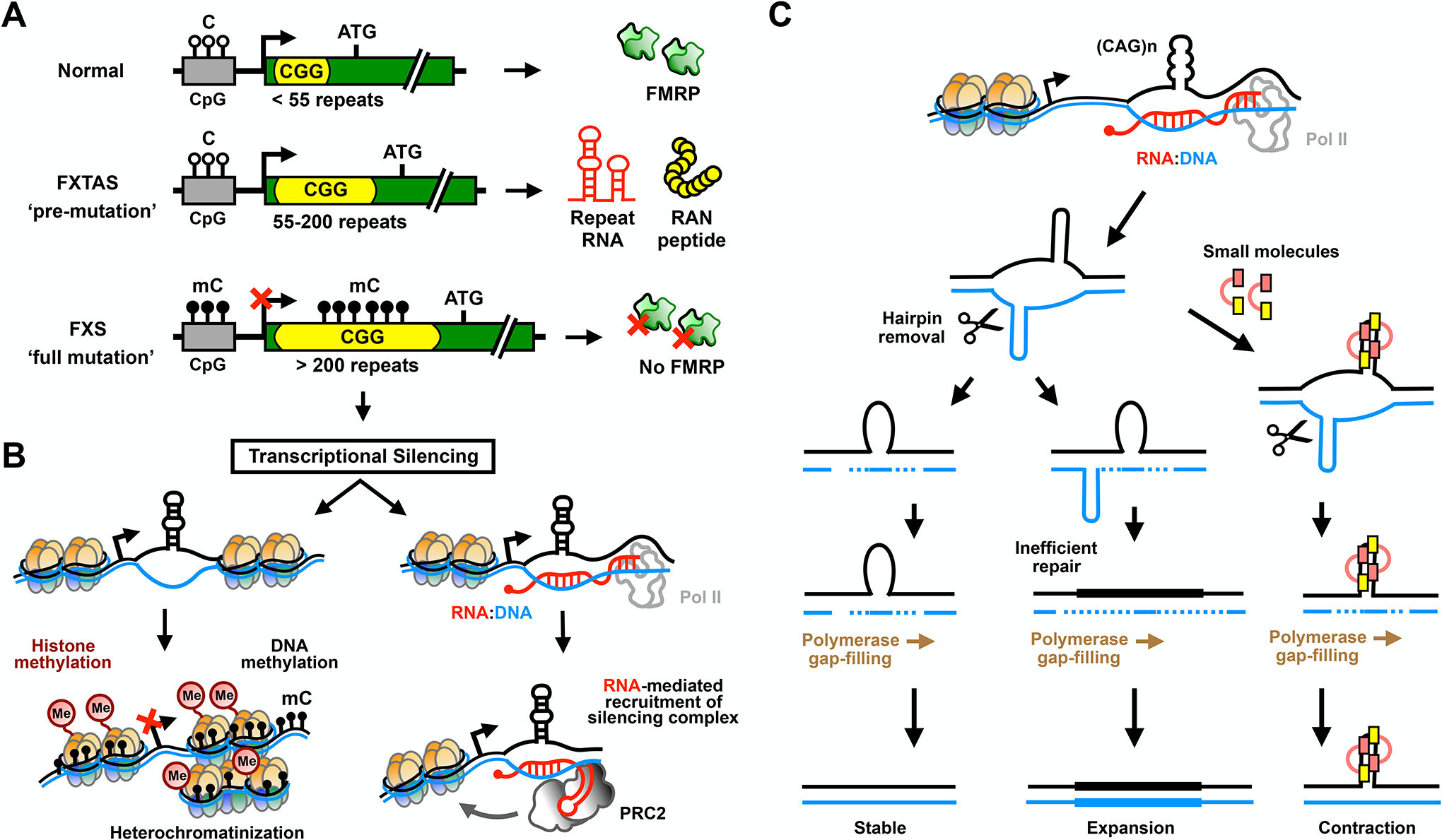
(A) Allelic classes of the FMR1 gene containing normal to pathogenic CGG repeats. FMR1 normally has ~30 CGG repeats in its 5’ UTR that are not included as a part of the mature protein product FMRP. Pre-mutation (55–200 repeats) expansions result in production of large CGG repeat-containing RNAs that underlie the age-related neurodegenerative disorder fragile X-associated tremor/ataxia syndrome (FXTAS). Full mutation (>200 repeats) expansions in subsequent generations lead to silencing of the FMR1 locus and fragile X syndrome (FXS). (B) CGG repeat methylation (mC) may direct transcriptional silencing by favoring histone methylation and heterochromatin formation through mechanisms similar to those typically active at CpG islands (left panel). Alternatively or cooperatively, nascent RNA may trigger epigenetic silencing by hybridizing to the complementary CGG-repeat DNA to form RNA:DNA duplexes that recruit polycomb repressive complex 2 (PRC2) (right panel). (C) Transcription-induced R-loops also support formation of DNA slip-out structures that contribute to repeat instability. For CAG/CTG trinucleotide repeat expansions, extended stable hairpins form in both strands. Normally, mismatch repair (MMR) pathways keep the repeat tract length stable by melting the slip-outs, followed by gap-filling by DNA polymerase across the region. Inefficient repair or formation of multiple slip-outs leads to retention of the slip-out structures and expansion of the repeat region by incorporation of the looped DNA. Small molecules that target slip-out structures in CAG repeat DNA inhibit repeat expansion and bias instability toward contraction.
