Figure 4: Mechanisms of repeat-associated non-AUG (RAN) translation.
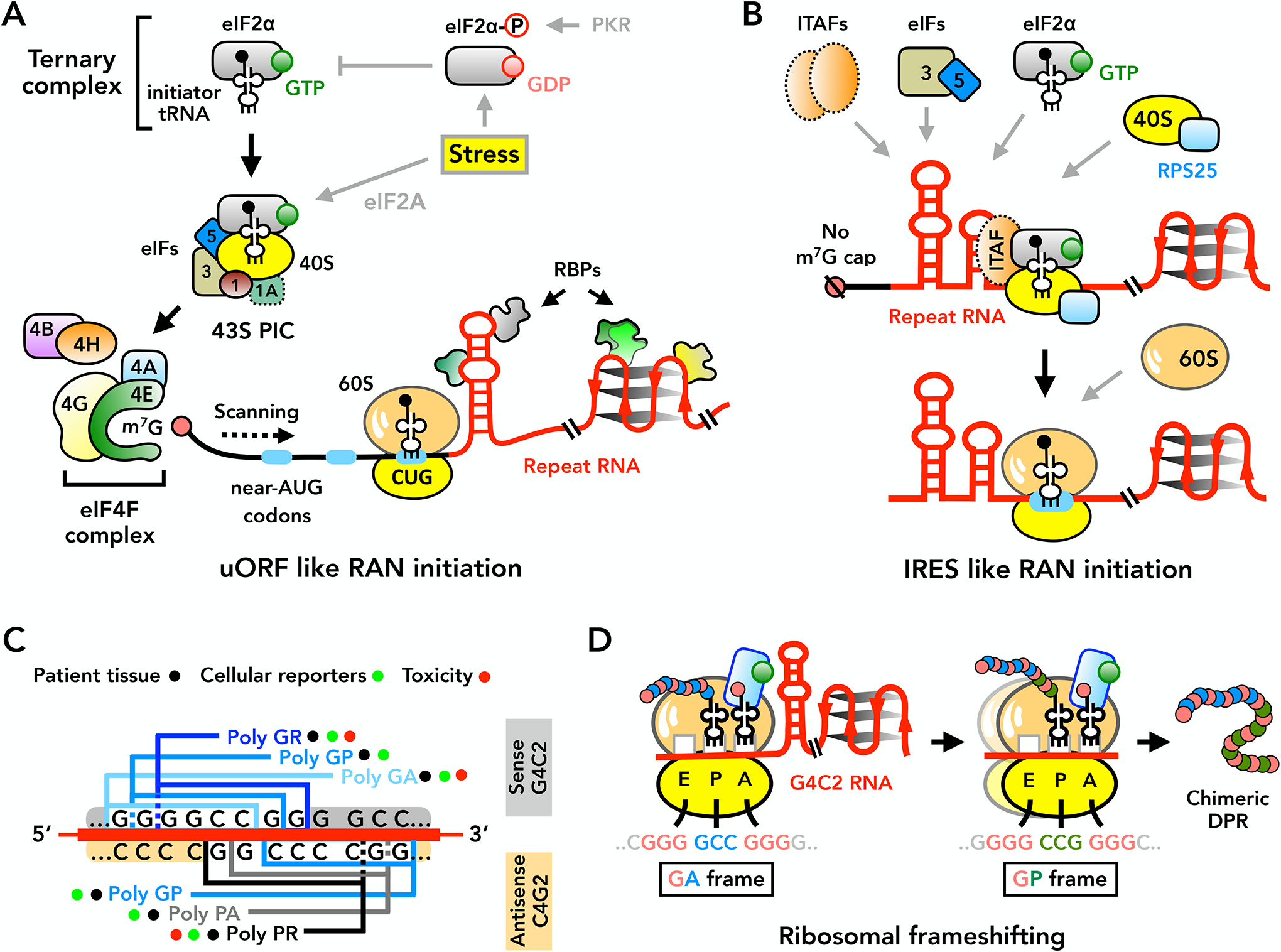
(A) Canonical AUG-mediated initiation and some forms of RAN translation require binding of eIF4F complex (eIF4E, eIF4G and eIF4A) to the 5’ m7G cap with eIF4B and/or eIF4H. After assembly, the 43S pre-initiation complex (PIC) scans 5’ to 3’ along the mRNA until selecting an AUG or near-AUG codon (for example: CUG) for initiation. eIF2α phosphorylation (eIF2α-P) under stress blocks ternary complex recycling and inhibits canonical translation, but allows for continued RAN translation. RBPs regulate RAN initiation by binding and altering repeat RNA structures. Known RAN-associated factors are depicted with solid lines, while canonical and IRES initiation factors involved in RAN translation are depicted with dashed lines. (B) RAN translation may also initiate through IRES-like mechanisms in a cap-independent manner, supported by RPS25 and other IRES-trans acting factors (ITAFs). (C) RAN translation from the C9ORF72 GGGGCC sense and CCCCGG antisense transcripts generates multiple dipeptide repeats (DPRs). While all DPRs are detected in patient tissues or generated by cellular reporters, arginine-containing DPRs show the highest intrinsic toxicity in model systems. (D) Stable RNA secondary structures formed by GGGGCC repeats induce ribosomal frameshifting during RAN translation, leading to production of chimeric DPRs. 40S and 60S = ribosomal subunits, eIF = eukaryotic initiation factor, IRES = internal ribosome entry site, m7G = 7-methylguanosine, PKR = Protein kinase R, RBPs = RNA-binding proteins, uORF = upstream open reading frame.
