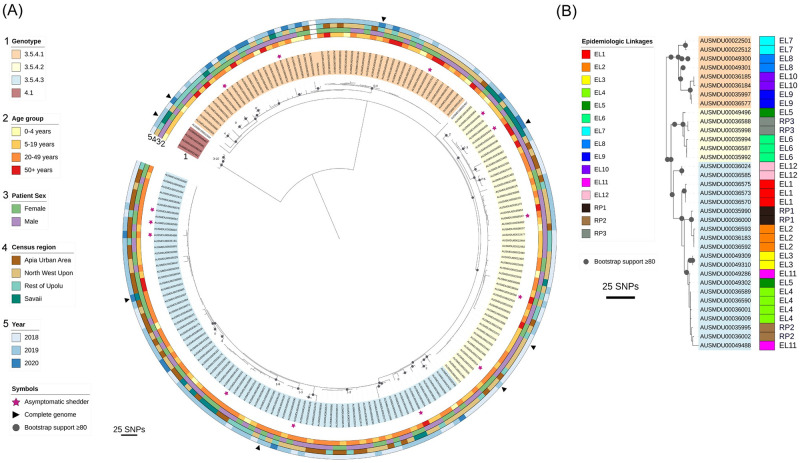
Fig 3. Maximum-likelihood phylogeny, associated epidemiologic variables, and epidemiologic linkages among S. Typhi from Samoa from April 2018 through June 2020.
(A) Maximum-likelihood phylogeny from de novo assembled whole genome sequences of Samoan S. Typhi genotype 3.5.4 isolates from April 27, 2018 through June 9, 2020 (N = 186, S1 Table). Core genome single nucleotide polymorphisms (SNPs) were identified relative to the 2012 Samoa Reference genome H12ESR00394-001A (GCA_001118185.2). Genotype/sub-lineage is indicated by isolate label shading (ring 1). Two outlier isolates are not shaded. Asymptomatic shedders of S. Typhi are denoted with a pink star. The patient age group (ring 2), patient sex (ring 3), census region of residence (ring 4), and year of isolation (ring 5) are annotated in the circumscribing colored rings. (B) Epidemiologic linkages (EL) identified through case household investigations are compared against genomic relatedness of the isolates based on core genome phylogeny. The visible isolates were extracted from the tree in Panel A. In total 2/12 epidemiologic linkages are not supported by the genomic variation observed in the isolates from those samples (EL5/dark green and EL11/pink groups are not similar). Three repeat positive (RP) blood cultures isolated ~1 month apart are also compared, showing close genetic similarity.