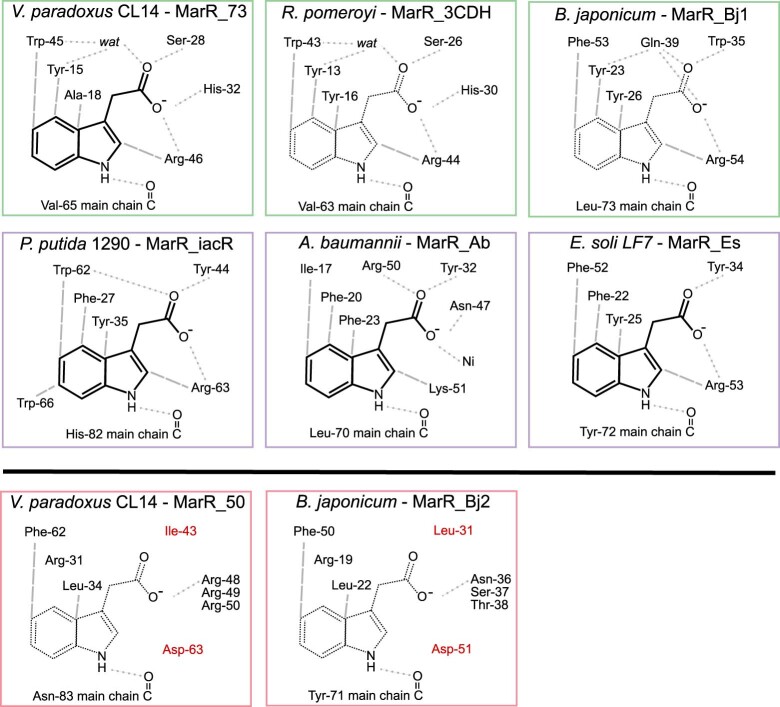
Extended Data Fig. 7. Key IAA-binding residues Conserved in MarR-family proteins.
At top, five crystal structures reported here along with the previously reported structure 3CDH; four of these structures contain bound indole-3-acetic acid (IAA, chemical structure shown as solid lines) and two were in unliganded states (IAA chemical structure shown as dashed). Hydrogen bonding interactions between protein groups and ligand are indicated with grey dotted lines, and non-hydrogen bonds with gray dashed lines. Outline colors correspond to Variovorax MarR_73-like (green), iac-like (purple), and Variovorax MarR_50-like (red) MarR types. The three top Variovorax MarR_73-like (green) proteins cluster together and exhibit highly similar ligand-binding residues. The three iac-like (purple) proteins, at middle, also cluster together and employ residues distinct from those at top, while maintaining similar contacts to IAA. The two Variovorax MarR_50-like (red) proteins, at bottom, do not have representative crystal structures; however, analysis of amino acid positions equivalent in sequence reveal that key contacting residues in the IAA-binding proteins at top are not conserved, specifically those highlighted in red.