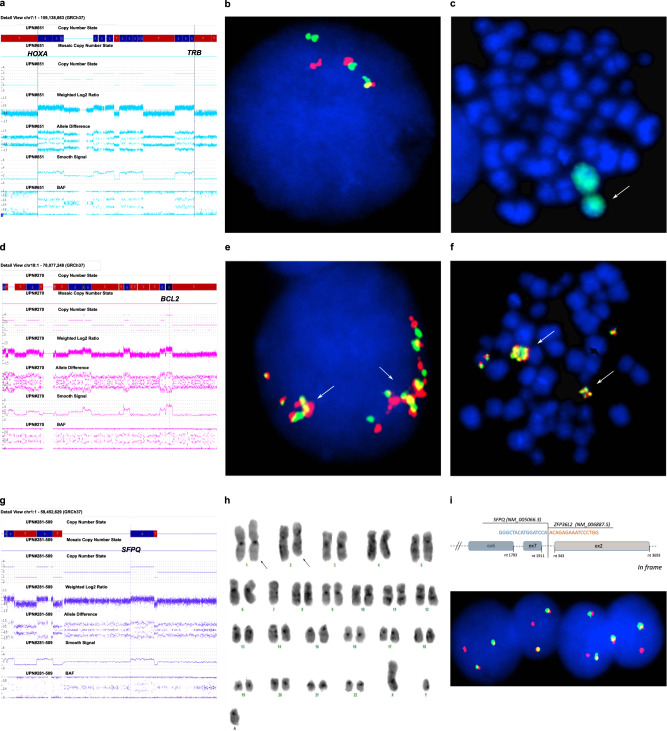
Fig. 1. Known and novel oncogenic events associated with chromothripsis.
Chromothripsis generated oncogenetic events in T-ALL: a SNPa profile of chromosome 7 showing breakpoints within the HOXA gene cluster and TRB@ (case no. 7, Table 1); b A double color double fusion FISH assay confirmed the TRB@::HOXA rearrangement in an interphase nucleus, i.e., 2 fusions, 1 orange and 1 green signal (case no. 7); c Metaphase FISH, with whole chromosome paint 7 (green), showed that one chromosome 7 was involved in a ring chromosome (white arrow) (case no. 7); d SNPa profile of chromosome 18 with a gain at BCL2 locus (case no. 8, Table 1); e, f FISH with the LSI BCL2 break-apart FISH probe (Vysis-Abbott, Milan, Italy) confirmed the gene was present as clusters of multiple fusion signals in interphase nuclei (e) and in an abnormal metaphase (f) (white arrows); g SNPa profile of chromosome 1 with a breakpoint at SFPQ (case no. 11, Table 1); h The G-banded karyotype of case no. 11 showing the reciprocal translocation between chromosomes 1 and 2 (black arrows); i The SFPQ-ZFP36L2 in frame fusion transcript detected by RNA sequencing (upper part); the rearrangement of ZFP36L2 was confirmed by the break-apart FISH assay with RP11–339H12 (orange) and RP11–391M15 (green) that gave 2 fusions and 1 orange signal) indicating a breakpoint occurred within clone RP11–339H12 (case no. 11).