Abstract
Background & Aims:
Early-onset colorectal cancer (EOCRC) is a distinct clinical and molecular entity with poor survival outcomes compared to late onset CRC (LOCRC). Although the incidence of EOCRC is rising, current CRC screening strategies have several limitations in diagnostic performance for EOCRC. In view of this clinical challenge, novel and robust biomarkers for detection of EOCRC are necessary. The aim of this study is to develop a circulating miRNA signature for the diagnosis of patients with EOCRC.
Methods:
A systematic discovery approach by analyzing a large, publicly available, noncoding RNA expression profiling dataset (GSE115513) was used. A panel of miRNAs was identified, which was subsequently validated in blood samples from EOCRC patients in two independent cohorts (n=149) compared with controls (n=110) and pre/post-operative plasma specimens (n=22) using qRT-PCR assays.
Results:
In the discovery phase, four miRNAs were found to be expressed in blood samples. A combination signature of these four miRNAs (miR-193a-5p, miR-210, miR-513a-5p and miR-628-3p) yielded an area under the curve (AUC) of 0.92 (95% CI=0.85-0.96) for identification of EOCRC in the training cohort. The miRNA panel performance was then confirmed in an independent validation cohort (AUC=0.88, 95% CI=0.82-0.93). Moreover, the miRNA panel robustly identified early stage EOCRC patients (P < 0.001). The decreased expression of miRNAs in post-surgery plasma specimens indicated their tumor-specificity.
Conclusions:
Our novel miRNA signature for the diagnosis of EOCRC has the potential to identify EOCRC patients with high accuracy for clinical application in the non-invasive diagnosis of EOCRC.
Keywords: miRNA panel, early onset colorectal cancer, diagnostic biomarker, liquid biopsy
Graphical Abstract
LAY SUMMARY:
Our blood-based circulating miRNA signature has the potential to identify patients with early onset colorectal cancer patients with high accuracy and offers a clinical tool for early diagnosis and population screening of patients suffering from this malignancy.
INTRODUCTION
Colorectal cancer (CRC) ranks third in cancer incidence and remains the third-leading cause of cancer-related deaths worldwide, in both men and women.1 In recent years, the incidence of sporadic CRC and associated mortality has declined worldwide.2 These improvements are likely due to the increased efforts in CRC screening that have allowed timely detection and removal of premalignant lesions and early-stage cancers, as well as improvements in treatment modalities.3–6 Unfortunately, data from epidemiological studies have revealed a significant trend for a rise in CRC incidence among individuals younger than 50 years of age, who do not possess any familial or hereditary disposition for this malignacy.1, 7, 8 This disease, referred to as the early-onset CRC (EOCRC), currently accounts for 10-15% of all new CRC diagnoses.6, 9, 10 At the current rate, it is estimated that the incidence for EOCRC is likely to double by the year 2030.7 While the reasons for this concerning trend in the younger population are poorly understood, there is a general consensus that patients with EOCRC are epidemiologically, biologically and pathologically distinct vis-à-vis those with late-onset colorectal cancer (LOCRC; patients ≥50 years old).11–15 Accordingly, for further characterization of EOCRC, these patients must be explored, assessed and clinically managed distinctly from those with LOCRC.
Notably, recent studies have suggested that patients with EOCRC have distinct clinical behaviors compared to those with LOCRC.16 EOCRC patients are more likely to exhibit an advanced stage tumor at initial presentation, distal tumor localization, signet ring histology, and a disease presentation with concurrent metastasis.17–20 This raises the logistical clinical concern that, since the tumors in EOCRC patients are often more aggressive than those with LOCRC, a delayed diagnosis could have a significant adverse impact and can lead to early death. Given the earlier age of onset and increased disease severity, these data highlight the need to develop screening approaches that can facilitate earlier detection and timely intervention for improving the overall survival in patients afflicted with EOCRC.
Recently, the guideline for initial CRC screening in the general population was lowered to 45 years by the American Cancer Society.21 However, this screening approach can benefit only a small fraction of younger individuals, as the overwhelming majority of these are likely not to have sporadic EOCRC, but are more likely to be those with a familial or hereditary form of CRC (e.g. Lynch syndrome). From a practical standpoint, lowering the age for CRC screening to capture patients with EOCRC using conventional colonoscopy has its challenges, as this procedure is invasive, has attendant risks, and is cost prohibitive for implementation in the average risk-population. In addition, currently available non-invasive tests including fecal and blood tests lack adequate diagnostic performance for the early detection of CRC,22–25 especially EOCRC, as these assays have yet to be explored or developed in this population. These limitations highlight the imperative need to develop robust, non-invasive biomarkers that can help overcome the challenges of the current generation of diagnostic assays, and facilitate the identification of patients with EOCRC.
Liquid biopsies have become highly topical in the field of diagnosis for various malignancies, and are gaining attention for overcoming some of the limitations of current screening strategies.26, 27 In particular, blood remains the most attractive substrate for developing liquid biopsy-based assays as it is widely accepted by patients, and carries cancer-derived cargo released into the systemic circulation including proteins, metabolites and nucleic acids.28 In this context, work from our group and others has previously shown that circulating microRNAs (miRNAs) reflect physiological and pathological alterations in patients with CRC,29 and have the potential to serve as important surrogates as minimally invasive diagnostic biomarkers.30, 31 MiRNAs are short, 18–24-nucleotide, non-coding RNAs that play a pivotal role in gene regulation. Since miRNA expression is generally stable in tissues, blood, stool, and other bodily fluids, they have emerged as promising candidates for developing liquid biopsy biomarkers in human cancers.32 While, miRNAs have been previously explored in diagnosis of LOCRC,33–36 to the best of our knowledge, no systematic studies have yet focused on identifying such biomarkers for the detection of patients with EOCRC.
Given the rise in incidence of EOCRC and the challenges for its early detection, we performed a systematic, genome-wide analysis to comprehensively identify a miRNA signature for the early detection of patients with EOCRC. Following the biomarker discovery, we rigorously validated this signature in multiple independent cohorts of patients, and finally established a novel liquid biopsy assay for the detection of patients with early-stage EOCRC.
MATERIALS AND METHODS
The present study employed a two-phase design. In the initial biomarker discovery phase, transcriptomic data from miRNA expression profiling dataset was systematically analyzed for the identification of a clinically translatable miRNAs that detects EOCRC. In the second phase, plasma specimens from multiple clinical cohorts were used to validate the performance of biomarkers selected in the discovery phase. In this clinical validation phase, quantitative reverse-transcription PCR (qRT-PCR) assays were employed to evaluate and compare the expression levels of candidate miRNAs in plasma specimens from individuals with EOCRC and those of no disease. The overall workflow for this study is illustrated in Figure 1A.
Figure 1.
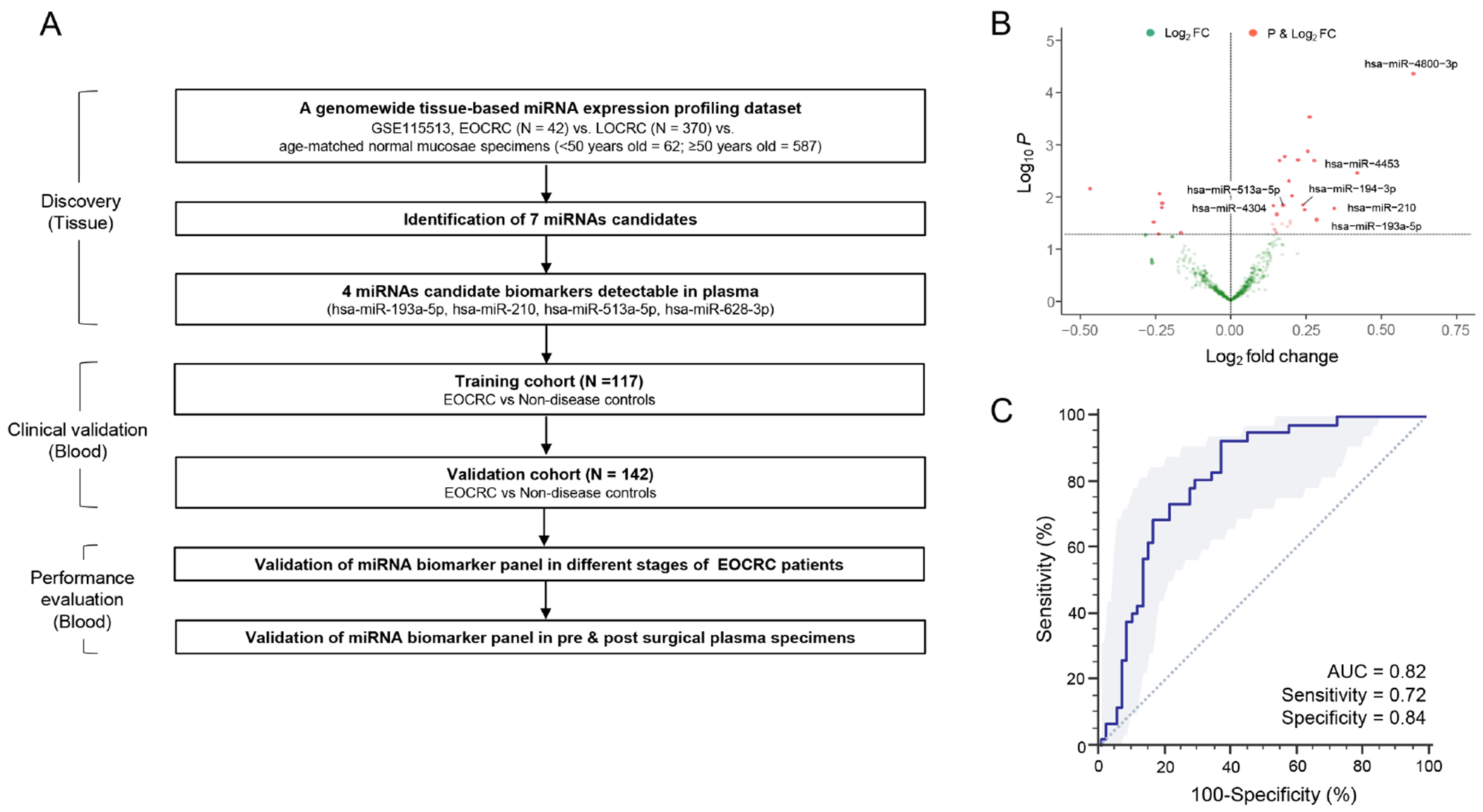
Differentially expressed miRNAs in patients with early stage EOCRC vs. normal mucosa specimens. (A) The overall workflow for this study. (B) A volcano plot illustrating miRNAs significantly upregulated in EOCRC (stage I-II) vs. normal mucosa specimens. (C) Receiver operating characteristics (ROC) curve analysis with the selected 7 candidate miRNAs for discriminating stage I/II EOCRC tumors. *P<0.05
Identification and selection of candidate miRNAs
A large, publicly available, miRNA expression profiling dataset (GSE115513, n=1061) was analyzed for the systematic discovery of miRNA biomarkers specific for patients with EOCRC. The normalized and pre-processed miRNA expression data was downloaded from the Gene Expression Omnibus (GEO). The miRNA expression data from paraffin-embedded tissues with patients with either stage I/II EOCRC (n = 42) or LOCRC (n = 370), as well as age-matched normal mucosa samples (<50 years old = 62; ≥50 years old = 587), respectively, were analyzed to identify candidate miRNAs, preferentially those that were upregulated in EOCRC. The differential expression analysis was performed using limma (version 3.38.3). Furthermore, the effect of any inadvertent potential bias that might exist due to age was mitigated by removing the miRNAs with a significant (P < 0.05) change in the expression in the normal samples stratified by age (<50 and ≥50 years of age). A multivariate logistic regression model was built to establish the diagnostic potential of upregulated miRNAs in patients with EOCRC. Multi-collinearity was evaluated by calculating a variance inflation factor (VIF) for each predictor. The VIF values were calculated for each parameter in the model, and if found higher than the cutoff (= 2), sequentially the predictor with the highest VIF was dropped, recalculated, and repeated until all values were below the cutoff.
Patient cohorts and specimens
For this study, a total of 349 plasma samples, which included 117 samples in the training cohort (72 from patients with EOCRC and 45 from healthy donors < 50 years old or non-disease controls) from 3 Japanese hospitals (National Cancer Center Hospital, Mie University, and Nagoya University), and 142 samples in the validation cohort (77 EOCRC and 65 non-disease controls) from Spanish hospitals (Hospital Universitario de Canarias, Hospital Clinic de Barcelona, Hospital Universitario de Donostia and Hospital Universitario Fundación Jiménez Díaz) were evaluated. For additional analyses, 44 specimens from patients with LOCRCs and 24 age-matched non-disease controls (≥50 years old), as well as a subset of 22 plasma specimens from patients that were obtained both pre- and post-operatively were used. Tumor staging was performed according to the Sixth Edition of the American Joint Commission on Cancer TNM staging system. The study was conducted in accordance with Declaration of Helsinki. Written informed consent was obtained from all patients, and the study was approved by the institutional review board of all participating institutions.
RNA extraction, cDNA synthesis and quantification of miRNAs using qRT-PCR
Total RNA was extracted from plasma samples using miRNeasy Serum/Plasma Kits (Qiagen, Valencia, CA; catalog number 217184) following manufacturer’s protocol. Briefly, 250 μL of plasma was thawed on ice and centrifuged at 16,000 × g at 4°C for 10 minutes to remove cellular debris. Thereafter, 200 μL of supernatant was lysed in 1000 μL of QIAzol Lysis Reagent. After incubation for 5 minutes, 25 fmol of synthetic cel-miR-39 (Syn-cel-miR-39-3p miScript miRNA Mimic, Qiagen; catalog number MSY0000010) was added to each sample as an external spiked-in control. Total RNA, including small RNA, was extracted and eluted in 30 μL of RNase-free water using a QIAcube (Qiagen). Complementary DNA (cDNA) from total RNA was synthesized following miRCURY LNA (Qiagen). First-Strand cDNA Synthesis protocol was employed with slight modifications. qRT-PCR analysis was performed using the SensiFAST™ probe Lo-ROX Kit (Bioline, London, UK) on the QuantStudio 7 Flex Real Time PCR System (Applied Biosystems, Foster City, CA), and miRNA expression levels were evaluated with Applied Biosystems QuantStudio 7 Flex Real Time PCR System Software. The relative abundance of target transcripts was evaluated and normalized against the control using the 2−ΔΔCt method. After the evaluation of different endogenous control miRNAs including U6, miR-16-5p and miR-103a-3p, miR-103a-3p was determined to be the optimal control based on expression and uniform threshold cycle (Ct) values across all samples (non-disease controls and cancer). Normalized values were further log10 transformed 37, 38.
Statistical analysis
The selection criteria to identify differentially expressed miRNAs included a log2 fold change higher than zero and a p-value of less than or equal to 0.05. The data pre-processing and handling was performed using R/Bioconductor. Multicollinearity was tested using vif function in “rms”, and R library. A generalized multivariate regression model was built. Receiver operator characteristic (ROC) curves, areas under curve (AUC), positive predictive values (PPV), and negative predictive values (NPV) were determined to measure the performance of the diagnostic model. Thereafter, the Youden’s index derived optimal cut-off thresholds were used to divide the training cohort patients with low vs. high-risk scores for diagnosing EOCRC. In order to analyze the miRNA model performance in different stages, we divided our cohort in stage I/II vs. III-IV EOCRC and analyzed its diagnostic performance. Statistical analyses for this study were performed using GraphPad Prism version 8.0, Medcalc statistical software V.16.2.0 (Medcalc Software bvba, Ostend, Belgium), and R (3.5.0, R Development Core Team, https://cran.r-project.org/). All p-values less than 0.05 were considered significant.
RESULTS
Genome-wide transcriptomic profiling identifies a 7-miRNA tissue-based signature for the detection of patients with EOCRC
The first objective of this project was to identify a systematic and comprehensive miRNA signature from genome-wide transcriptomic profiling data available for patients with early-stage EOCRC compared to those with LOCRC and age-matched controls. In these analyses, 28 miRNAs were significantly up-regulated in patients with stage I-II EOCRC tissue samples compared to the normal mucosae (P < 0.05). To enhance the overall specificity of the discovered markers, all miRNAs that overlapped between EOCRC and LOCRC patients were removed to increase the diagnostic potential of the markers that preferentially allowed identification of patients with EOCRC. Such a bioinformatic prioritization effort resulted in a panel of 11 miRNAs that were significantly upregulated in patients with EOCRC (P < 0.05). Subsequently, after removing the biomarkers that exhibited collinearity with each other and did not offer any additional diagnostic contribution to the marker panel, a final panel of seven miRNAs that represented up-regulated markers in EOCRC patients (Fig.1B) was prioritized. Finally a multivariate logistic regression model established the association of these seven miRNAs (hsa-miR-4304, hsa-miR-513a-5p, hsa-miR-628-3p, hsa-miR-194-3p, hsa-miR-193a-5p, hsa-miR-210 and hsa-miR-4453) in patients with stage I/II EOCRC, which exhibited a diagnostic AUC value of 0.82 (95% CI=0.73-0.91, P < 0.001), with a corresponding a sensitivity of 0.72 and specificity of 0.84 for the detection of patients with early stage EOCRC (Fig.1C) The data highlighted that this biomarker panel possessed remarkable diagnostic potential for the diagnosis of patients with EOCRC.
Establishment and validation of diagnostic performance of a circulating miRNA panel in independent cohorts of patients with EOCRC
Since the primary objective of our project was to develop a minimally invasive, liquid biopsy based assay for the identification of patients with EOCRC in clinical settings, the validation of diagnostic performance of this miRNA panel in actual clinical settings was warranted. In order to develop a blood-based assay, the tissue-based markers identified during the biomarker discovery phase were tested for detectability in plasma samples. Four of the seven tissue-based markers (hsa-miR-513a-5p, hsa-miR-628-3p, hsa-miR-193a-5p and hsa-miR-210) were detectable in these plasma samples. Regarding the functional relevance of the miRNAs, they serve as transcripts of several key genes associated with pathogenesis in cancer. The details pertaining to these differentially expressed miRNAs, the corresponding genes and their clinical significance are presented in Supplementary Tables 1 and 2.
Subsequently, to establish the diagnostic potential of this miRNA panel for EOCRC patients, its performance in blood specimens obtained from two large, independent clinical cohorts was examined by using quantitative qRT-PCR assays. The clinical features of the patient cohort are summarized in Table 1. In these experiments, using logistic regression analysis, the performance of this biomarker panel was trained in a cohort of patients from Japan [n = 117 (72 EOCRC and 45 non-disease controls)]. The miRNA expression-based risk scores for each patient with EOCRC were calculated as follows: Logit (P) = (−8.78655*MIR193) + (1.77237*MIR210) + (5.30584*MIR513) + (1.65455*MIR628) + 3.75375. This logistic regression based four miRNA risk-assessment model demonstrated an excellent diagnostic performance in blood specimens of patients with EOCRC with an AUC value of 0.92 (95% CI = 0.85-0.96, P < 0.001; Fig.2A), with a corresponding sensitivity of 0.90, specificity of 0.80, PPV of 0.88 and NPV of 0.84 (Table 2).
Table 1:
Clinicopathologic characteristics of the training and validation cohorts.
| Training cohort (N =117) | Validation cohort (N = 142) | |||
|---|---|---|---|---|
|
| ||||
| EOCRC (N = 72) |
Controls (N = 45) |
EOCRC (N = 77) |
Controls (N = 65) |
|
| Age (median, range) | 44 (21-49) | 37 (26-49) | 43 (24-49) | 45 (39-49) |
| Gender (n, %) | ||||
| Male | 32 (44.4%) | 27 (60.0%) | 37 (48.1%) | 33 (50.8%) |
| Female | 40 (55.6%) | 18 (40.0%) | 40 (51.9%) | 32 (49.2%) |
| Localization | ||||
| Right side | 19 (26.4%) | - | 17 (22.1%) | - |
| Left side | 36 (50.0%) | - | 32 (41.6%) | - |
| Rectum | 17 (23.6%) | - | 28 (22.1%) | - |
| Stage (TNM) | ||||
| I | 20 (27.8%) | - | 9 (11.7%) | - |
| II | 19 (26.4%) | - | 19 (24.7%) | - |
| III | 25 (34.7%) | - | 29 (37.7%) | - |
| IV | 8 (11.1%) | - | 20 (26.0%) | - |
EOCRC: Early-onset colorectal cancer
Figure 2.
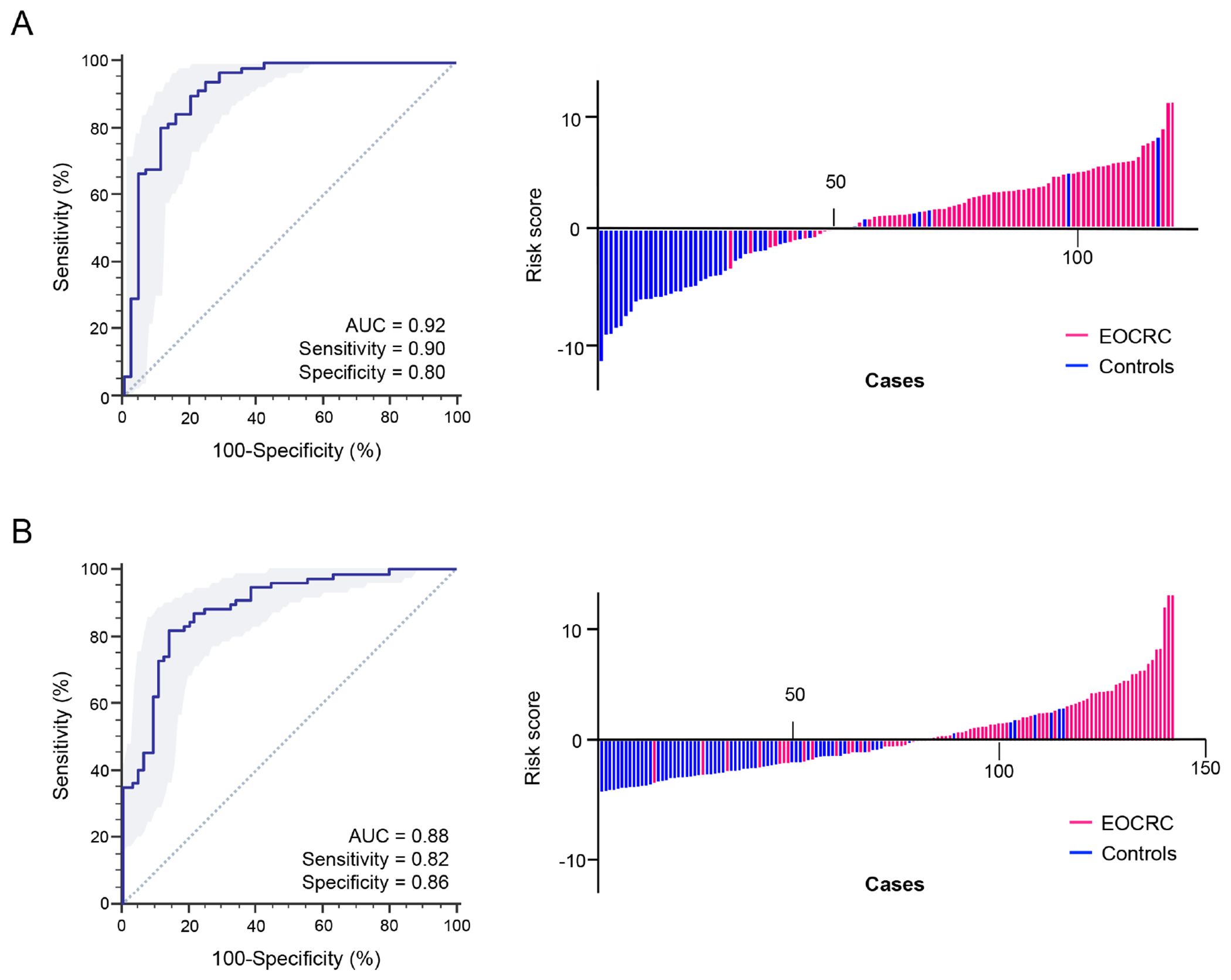
Diagnostic performance of the miRNA panel in EOCRC vs. non-disease controls. (A) The ROC curves analysis for a 4-miRNA panel in the training cohort. (B) Risk score distribution plot in the training cohort. (C) ROC curves analysis for a 4-miRNA panel in the validation cohort. (D) Risk score distribution plot in the validation cohort.
Table 2:
Summary of diagnostic performance of miRNA-based biomarker panel in the training and validation cohorts.
| Training cohort (%, CI) | Validation cohort (%, CI) | |
|---|---|---|
| AUC | 0.92 (0.85-0.96) | 0.88 (0.82-0.93) |
| Accuracy | 0.86 (0.79-0.92) | 0.84 (0.77-0.89) |
| Sensitivity | 0.90 (0.81-0.96) | 0.82 (0.71-0.90) |
| Specificity | 0.80 (0.65-0.90) | 0.86 (0.75-0.94) |
| PPV | 0.88 (0.78-0.94) | 0.88 (0.78-0.94) |
| NPV | 0.84 (0.69-0.93) | 0.80 (0.69-0.89) |
AUC, area under the curve; PPV, positive predictive value; NPV, negative predictive value
In view of the encouraging results of the blood-based panel for the detection of EOCRC in the training cohort, the robustness and accuracy of our risk-assessment model were assessed in another large independent validation cohort from Spain [n = 142 (77 EOCRC and 65 non-disease controls)]. These validation efforts confirmed the earlier findings and yielded an AUC value of 0.88 (95% CI = 0.82-0.93, P < 0.001; Fig.2B) with a sensitivity of 0.82, a specificity of 0.86, PPV of 0.88 and NPV of 0.80. Taken together, the genome-wide transcriptomic profiling approach was indeed robust, as it identified the biomarkers that were successfully trained and validated in plasma specimens from independent cohorts of patients with EOCRC, hence highlighting their translational potential in the clinic for the detection of this malignancy in early stages.
The circulating miRNA panel identifies early stage (stage I and II) and late stage (stage III and IV) EOCRC patients
One approach for improving the survival outcomes in patients with EOCRC is to attain an earlier diagnosis. Therefore, whether this biomarker panel performed better in subgroups of EOCRC patients based upon their tumor stage, i.e., early stage (Stage I and II) vs late stage (Stage III and IV) was questioned. It was exciting to observe that, in the validation cohort, this circulating biomarker panel revealed excellent diagnostic performance in the identification of early-stage cancers (stage I/II) with an AUC of 0.92 (95% CI 0.84-0.96, sensitivity 0.92, specificity 0.80). On the other hand, regarding late-stage cancers (stage III/IV), the miRNA panel showed relatively fair performance as well, with an AUC of 0.87 (95% CI 0.79-0.92, sensitivity 0.79, specificity 0.86; Fig.3A). Furthermore, the distribution of risk scores in early stage vs. late stage EOCRC patients relative to the non-disease controls was evaluated. It was quite reassuring to observe that this circulating biomarker panel successfully identified patients with both early and late stage EOCRCs in the validation cohort (P < 0.001, respectively; Fig.3B). Collectively, these results highlight that this four-miRNA circulating biomarker panel is robust in the identification of patients with early disease stages and late disease stages. These data are quite encouraging since early detection is surely essential to improve the survival outcomes in patients with EOCRC.
Figure 3.
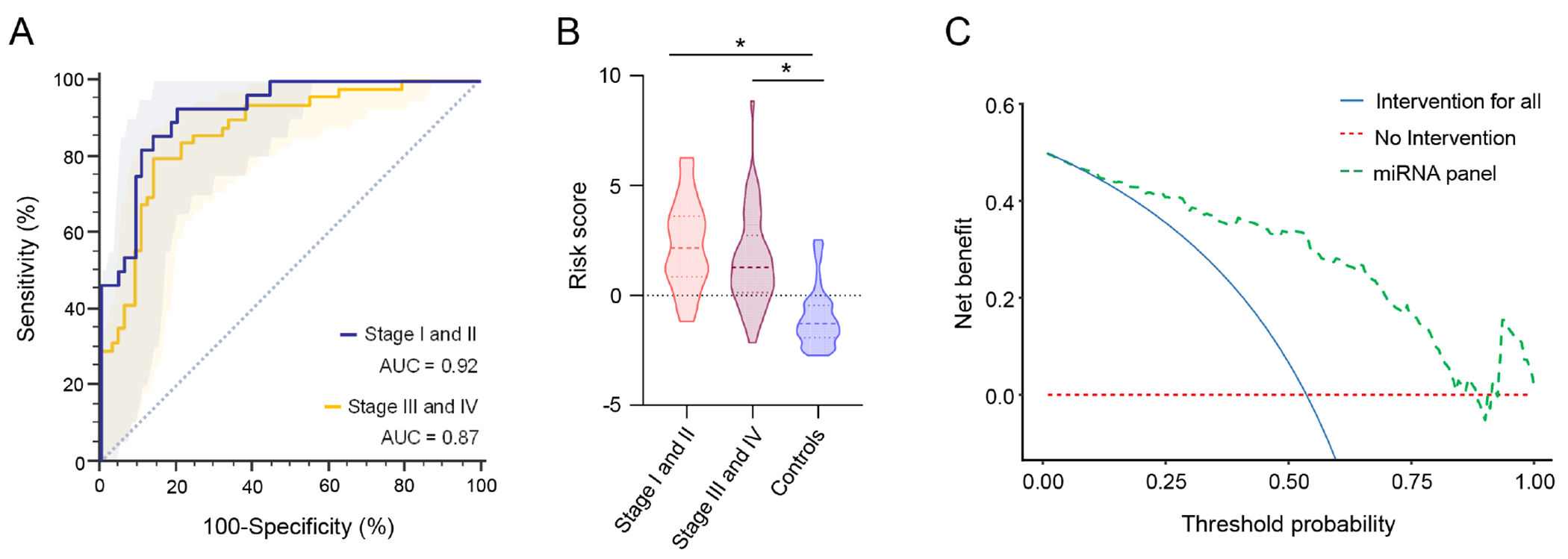
Diagnostic potential evaluation of the miRNA biomarker panel to identify different stages of EOCRC patients. (A) ROC curve analysis to identify early stage (I and II) and late stage (III and IV) patients with EOCRC from non-disease controls in the validation cohort and (B) Risk score analysis based on risk prediction formulae in early vs. late stage EOCRC patients and non-disease control in the validation cohort. (C) Decision curve analysis to evaluate the performance of the miRNA panel.
The miRNA panel offers a significant benefit compared to current screening approaches The clinical usefulness of screening strategies should be estimated by the trade-off between the harm and diagnosis. Since the diagnosis of CRC patients is achieved by a screening test through colonoscopy and an invasive biopsy, false positive or false negative cases would be detrimental to individuals undergoing such screening. To estimate the clinical significance of the miRNA panel, decision curve analysis (DCA) was performed (Fig. 3C). The DCA curve revealed that the miRNA panel achieved a higher net benefit regardless of threshold probability in comparison to intervention for all patients or none of the patients. These findings suggest that this miRNA panel might offer more clinical benefit with regards to the avoidance of physical harm and misdiagnosis.
The expression levels of circulating miRNAs significantly decrease following surgical removal of the CRC
The hypothesis behind the expression of circulating biomarkers is that tumor cells constantly shed cellular cargo into the systemic circulation, which is amenable to detection in a liquid biopsy. Presuming that the expression of these biomarkers diminishes once their source (i.e., the cancer) is resected, the expression of the miRNA biomarker panel was analyzed in a subset of EOCRC patients where the baseline plasma specimens were collected prior to surgery (pre-operative, n=12), and compared to the expression levels of these markers in the same patient’s plasma collected three months after curative surgery (post-operative; n=10). It was again reassuring to witness that the expression of each individual miRNA marker exhibited a statistically significantly decrease in expression in the post-operative blood specimens (P < 0.05; Fig.4A). Finally, in comparison with the risk probability in this subset of patients, these findings were again validated as a significantly reduced risk probability in post-operative vs pre-operative blood specimens was observed (P < 0.05; Fig.4B). These findings indicate that once the EOCRC was surgically resected, the levels of these biomarkers significantly decreased in the systemic circulation in the postoperative blood specimens. Taken together, these findings suggested that the four-miRNA panel is specific for EOCRC and may also have clinical potential as a disease-monitoring assay in future.
Figure 4.
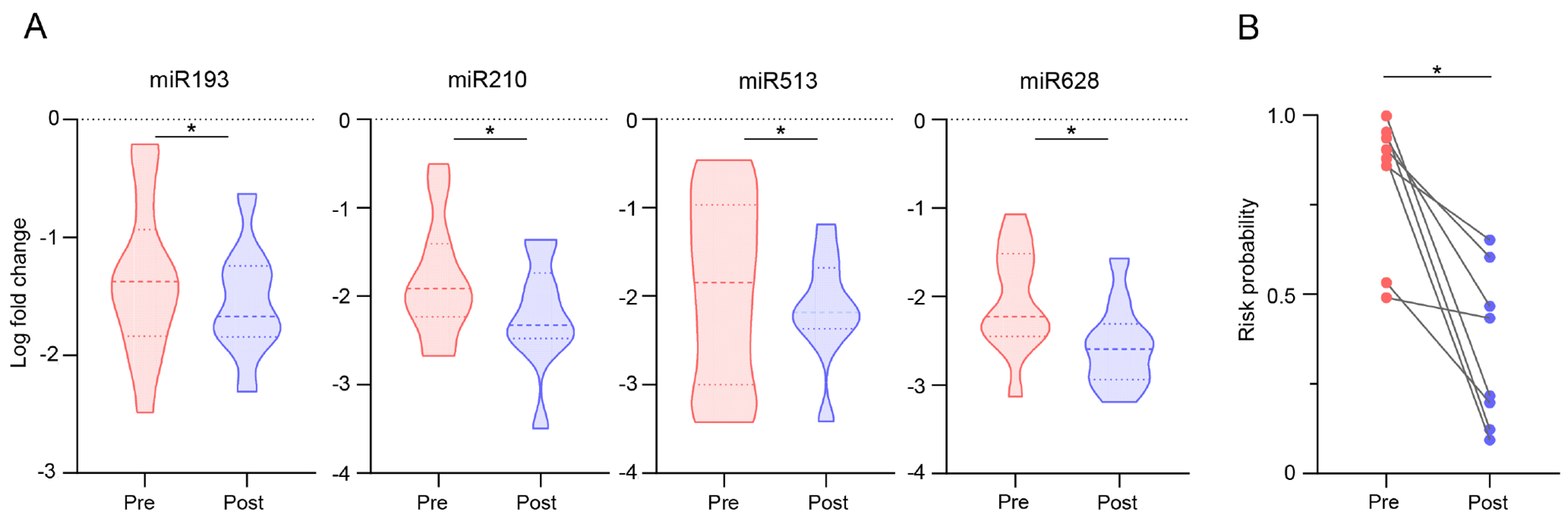
Evaluation of the miRNA panel in pre vs. post-operative blood specimens. (A) Expression of candidate miRNAs in pre- and post-operative plasma specimens from EOCRC patients. (B) Assessment of risk probability based on risk prediction formula between pre-and postoperative EOCRC specimens.
The circulating miRNA panel has the potential to identify patients with LOCRC
Considering that the age is the key discriminator for patients with EOCRC and LOCRC, the performance of miRNA panel was also evaluated among patients with LOCRC and compared with the age-matched non-disease subjects (≥50 years old). Interestingly, it was observed that the miRNA panel can also discriminate patients with LOCRC from age-matched non-disease controls with an AUC of 0.84 (95% CI 0.67-0.89, sensitivity 0.82, specificity 0.88; Supplementary Fig. 1). Collectively, these findings suggested that this circulating biomarker panel also has the potential to discriminate individuals with LOCRC from those with no disease.
DISCUSSION
In contrast to the decreasing trend in CRC-related mortality in adults 50 years or older, the overall incidence and mortality of EOCRC have been steadily rising.39, 40 To add to this clinical challenge, EOCRC cases are often detected at a more advanced stage, which highlights the need for their early detection in order to improve the overall survival in patients with this disease.6 Although colonoscopy remains the gold standard for the screening and diagnosis of CRC, it has major disadvantages including its invasive nature, potential risk of complications, and high costs. Furthermore, colonoscopy screening in the United States is often recommended at age 45, which will likely miss diagnosis of a large majority of patients with EOCRC. In view of these limitations, diagnostic approaches that are geared towards the early detection of patients with EOCRC remain an important unmet clinical need. In this effort to implement early detection strategies for EOCRC, the diagnostic modality should preferably be acceptable to healthy individuals, inexpensive, rapid and preferably non-invasive. Our present study is a significant step forward in this direction, where a systematic and comprehensive biomarker discovery effort has been undertaken, which led us to identify and establish a circulating miRNA signature that is highly robust in the identification of patients with EOCRC. Furthermore, the evidence that the expression of the discovered biomarkers was significantly reduced in plasma samples from patients in post-surgical settings was provided, hence suggesting that the tumor was indeed the source of these circulating markers in patients with EOCRC.
Currently, several types of non-invasive tests for CRC detection are available for clinical practice.22, 24, 25, 41 Stool tests, including fecal occult blood tests (FOBT), are commonly used for CRC screening.24 However, due to the poor sensitivity of older-generation guaiac-based fecal tests for advanced adenomas and cancer, the efficacy of these tests is limited,24 and increasingly those assays are being replaced by fecal immunochemical tests (FITs) for CRC screening in average risk populations.25 While the sensitivity of FIT for CRC varies widely (depending upon the cut-off value for the quantitative result),25, 42 some studies have demonstrated a sensitivity for CRC detection to range between 74-79%.43, 44 Although FITs is an attractive strategy for current noninvasive CRC screening, in order to overcome the limitations and improve its overall diagnostic accuracy, there is a need to develop additional diagnostic options that support current screening strategies. Our study is a significant step forward in this direction, where a four-miRNA panel in blood is reported, which is quite robust for the identification of patients with Stage I/II EOCRC. These findings highlight that our miRNA panel has the potential to improve the diagnostic performance of CRC screening that can be complemented with current clinical practice including FITs and might serve as an important non-invasive assay for the identification of patients with EOCRC.
Several previous studies have suggested that EOCRC is a biologically and clinically distinct disease compared to LOCRC.6, 40 Current CRC screening strategies mainly focus on LOCRC screening. Interestingly, our miRNA panel was also able to discriminate patients with LOCRC from age-matched non-disease controls with robust AUC values. A possible reason for this is that the older age cohort (≥50 years old) can include relatively younger individuals (early 50s) with CRC who might have developed their disease in their 40s that were potentially EOCRC and would have to be defined as EOCRC patients. Accordingly, our four-miRNA panel has potential to discriminate not only EOCRC patients (<50 years old) in the younger age cohort, but also potentially EOCRC patients (≥50 years old, defined as LOCRC, but developed in their 40s) in the older age cohort. These findings suggested that our miRNA panel may serve as an important biomarker for CRC screening that may be complemented with current LOCRC-focused strategy.
Several studies reported that four miRNAs in our miRNA panel (miR-193a-5p,45, 46 miR-21047–54, miR-513a-5p55–58 and miR-628a-3p59, 60) were overexpressed in cancer specimens and their high expression was correlated with poor survival or metastasis in different cancers. Several studies described that miR-210 overexpression is correlate with hypoxia and poor prognosis.47–49 In vitro and in vivo studies reported that overexpression of miR-193a-5p contributed to metastasis in pancreatic cancer.45 miR-513a-5p overexpression is associated with cancer metastasis56, 57 and can be a risk factor for breast cancer.58 miR-628a-3p were found to display significantly different expression levels in pancreatic cancer.59 A previous study reported that the expression level of miR-628a-3p was higher in cancer plasma samples in lung cancer, which might serve as liquid biopsy marker.60
It is important to acknowledge a few potential limitations of our study. Perhaps, the most important limitation of our study is the sample size of our patient cohorts, which was relatively modest. Therefore, further prospective studies using larger patient cohorts should be performed to successfully translate our findings to the clinic. Other potential limitations include, the availability of matched pre- and post-operative blood specimens from a larger cohort of patients with EOCRC. Moreover, our study cohort did not include patients with adenoma, thus further study is required to elucidate the performance of miRNA panel to diagnose patients with colorectal adenomas. Nevertheless, our work provides important evidence in determining the potential diagnostic capability of miRNAs for EOCRC, which could pave the way towards availability of robust molecular biomarkers for the risk-assessment and management of EOCRC patients.
In conclusion, using a systemic discovery approach, we have identified and developed, for the first time, a novel miRNA signature for the detection of EOCRC patients. The miRNA panel was successfully validated in two independent patient cohorts and robustly distinguished EOCRC patients from non-disease control samples. We envisage that pending further validation in much larger, future prospective studies, this four-miRNA biomarker panel has the potential to transform the screening of EOCRC patients and subsequently reduce the mortality rates.
Supplementary Material
WHAT YOU NEED TO KNOW.
BACKGROUND AND CONTEXT:
Although the incidence of early onset colorectal cancer is rapidly rising, there is a clear lack of screening strategies for the early detection of this disease.
NEW FINDINGS:
We have established a circulating microRNA signature that has the potential to identify patients with early onset colorectal cancer robustly and has the potential for use a noninvasive assay for population screening.
LIMITATIONS:
The sample size of our patient cohorts was relatively modest; hence, further prospective studies using larger patient cohorts might be needed for further validation and translation of our findings to the clinic.
IMPACT:
Our four-microRNA signature has the potential to transform the clinical practice by allowing non-invasive and timely detection of patients with early onset colorectal cancer.
ACKNOWLEDGEMENTS:
We would like to thank Drs. Souvick Roy, Satoshi Nishiwada, Tatsuhiko Kakisaka, Yasuyuki Okada, In-Seob Lee, Divya Sahu, Yinghui Zhao for their thoughtful discussions and input into the study. We also would like to extend our thanks to Dr. Sarah Wilkinson for her editorial assistance in the preparation of this manuscript.
Funding:
This work was supported by CA72851, CA181572, CA184792, CA202797, and CA227602 grants from the National Cancer Institute, National Institutes of Health. Goretti Hernández received a grant from Fundación Mapfre Guanarterme (Canarias, Spain) to work in this project.
Abbreviations:
- AUC
Area under the curve
- CRC
Colorectal cancer
- EOCRC
Early-onset colorectal cancer
- FIT
Fecal immunochemical test
- FOBT
Fecal occult blood test
- LOCRC
Late-onset colorectal cancer
- miRNA
Micro RNA
- NPV
Negative predictive value
- OR
Odds ratio
- PPV
Positive predictive value
- qRT-PCR
Quantitative reverse transcription polymerase chain reaction
- TNM
Tumor-node-metastasis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: None of the authors has any potential conflicts to disclose.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Malvezzi M, Carioli G, Bertuccio P, et al. European cancer mortality predictions for the year 2018 with focus on colorectal cancer. Ann Oncol 2018;29:1016–1022. [DOI] [PubMed] [Google Scholar]
- 3.Holme Ø, Bretthauer M, Fretheim A, et al. Flexible sigmoidoscopy versus faecal occult blood testing for colorectal cancer screening in asymptomatic individuals. Cochrane Database Syst Rev 2013:Cd009259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Fedewa SA, Anderson WF, et al. Colorectal Cancer Incidence Patterns in the United States, 1974-2013. J Natl Cancer Inst 2017;109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welch HG, Robertson DJ. Colorectal Cancer on the Decline--Why Screening Can’t Explain It All. N Engl J Med 2016;374:1605–7. [DOI] [PubMed] [Google Scholar]
- 6.Stoffel EM, Murphy CC. Epidemiology and Mechanisms of the Increasing Incidence of Colon and Rectal Cancers in Young Adults. Gastroenterology 2020;158:341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey CE, Hu CY, You YN, et al. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975-2010. JAMA Surg 2015;150:17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu PH, Wu K, Ng K, et al. Association of Obesity With Risk of Early-Onset Colorectal Cancer Among Women. JAMA Oncol 2019;5:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnett-Hartman AN, Lee JK, Demb J, et al. An Update on the Epidemiology, Molecular Characterization, Diagnosis, and Screening Strategies for Early-Onset Colorectal Cancer. Gastroenterology 2021;160:1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70:145–164. [DOI] [PubMed] [Google Scholar]
- 11.Kothari N, Teer JK, Abbott AM, et al. Increased incidence of FBXW7 and POLE proofreading domain mutations in young adult colorectal cancers. Cancer 2016;122:2828–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearlman R, Frankel WL, Swanson B, et al. Prevalence and Spectrum of Germline Cancer Susceptibility Gene Mutations Among Patients With Early-Onset Colorectal Cancer. JAMA Oncol 2017;3:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirzin S, Marisa L, Guimbaud R, et al. Sporadic early-onset colorectal cancer is a specific sub-type of cancer: a morphological, molecular and genetics study. PLoS One 2014;9:e103159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavestro GM, Mannucci A, Zuppardo RA, et al. Early onset sporadic colorectal cancer: Worrisome trends and oncogenic features. Dig Liver Dis 2018;50:521–532. [DOI] [PubMed] [Google Scholar]
- 15.Connell LC, Mota JM, Braghiroli MI, et al. The Rising Incidence of Younger Patients With Colorectal Cancer: Questions About Screening, Biology, and Treatment. Curr Treat Options Oncol 2017;18:23. [DOI] [PubMed] [Google Scholar]
- 16.Zaborowski AM, Abdile A, Adamina M, et al. Characteristics of Early-Onset vs Late-Onset Colorectal Cancer: A Review. JAMA Surg 2021;156:865–874. [DOI] [PubMed] [Google Scholar]
- 17.Aherne ST, Madden SF, Hughes DJ, et al. Circulating miRNAs miR-34a and miR-150 associated with colorectal cancer progression. BMC Cancer 2015;15:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yantiss RK, Goodarzi M, Zhou XK, et al. Clinical, pathologic, and molecular features of early-onset colorectal carcinoma. Am J Surg Pathol 2009;33:572–82. [DOI] [PubMed] [Google Scholar]
- 19.Goel A, Nagasaka T, Spiegel J, et al. Low frequency of Lynch syndrome among young patients with non-familial colorectal cancer. Clin Gastroenterol Hepatol 2010;8:966–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang DT, Pai RK, Rybicki LA, et al. Clinicopathologic and molecular features of sporadic early-onset colorectal adenocarcinoma: an adenocarcinoma with frequent signet ring cell differentiation, rectal and sigmoid involvement, and adverse morphologic features. Mod Pathol 2012;25:1128–39. [DOI] [PubMed] [Google Scholar]
- 21.Wolf AMD, Fontham ETH, Church TR, et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin 2018;68:250–281. [DOI] [PubMed] [Google Scholar]
- 22.Sørensen CG, Karlsson WK, Pommergaard HC, et al. The diagnostic accuracy of carcinoembryonic antigen to detect colorectal cancer recurrence - A systematic review. Int J Surg 2016;25:134–44. [DOI] [PubMed] [Google Scholar]
- 23.Su BB, Shi H, Wan J. Role of serum carcinoembryonic antigen in the detection of colorectal cancer before and after surgical resection. World J Gastroenterol 2012;18:2121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee MW, Pourmorady JS, Laine L. Use of Fecal Occult Blood Testing as a Diagnostic Tool for Clinical Indications: A Systematic Review and Meta-Analysis. Am J Gastroenterol 2020;115:662–670. [DOI] [PubMed] [Google Scholar]
- 25.Lee JK, Liles EG, Bent S, et al. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann Intern Med 2014;160:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heitzer E, Haque IS, Roberts CES, et al. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet 2019;20:71–88. [DOI] [PubMed] [Google Scholar]
- 27.Siravegna G, Mussolin B, Venesio T, et al. How liquid biopsies can change clinical practice in oncology. Ann Oncol 2019;30:1580–1590. [DOI] [PubMed] [Google Scholar]
- 28.Shigeyasu K, Toden S, Zumwalt TJ, et al. Emerging Role of MicroRNAs as Liquid Biopsy Biomarkers in Gastrointestinal Cancers. Clin Cancer Res 2017;23:2391–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hur K, Toiyama Y, Okugawa Y, et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 2017;66:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozawa T, Kandimalla R, Gao F, et al. A MicroRNA Signature Associated With Metastasis of T1 Colorectal Cancers to Lymph Nodes. Gastroenterology 2018;154:844–848.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wada Y, Shimada M, Murano T, et al. A Liquid Biopsy Assay for Noninvasive Identification of Lymph Node Metastases in T1 Colorectal Cancer. Gastroenterology 2021;161:151–162.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- 33.Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 2009;58:1375–81. [DOI] [PubMed] [Google Scholar]
- 34.Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 2010;127:118–26. [DOI] [PubMed] [Google Scholar]
- 35.Toiyama Y, Takahashi M, Hur K, et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst 2013;105:849–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zanutto S, Ciniselli CM, Belfiore A, et al. Plasma miRNA-based signatures in CRC screening programs. Int J Cancer 2020;146:1164–1173. [DOI] [PubMed] [Google Scholar]
- 37.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. Rna 2008;14:844–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kandimalla R, Gao F, Matsuyama T, et al. Genome-wide Discovery and Identification of a Novel miRNA Signature for Recurrence Prediction in Stage II and III Colorectal Cancer. Clin Cancer Res 2018;24:3867–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Low EE, Demb J, Liu L, et al. Risk Factors for Early-Onset Colorectal Cancer. Gastroenterology 2020;159:492–501.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akimoto N, Ugai T, Zhong R, et al. Rising incidence of early-onset colorectal cancer - a call to action. Nat Rev Clin Oncol 2021;18:230–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cross AJ, Wooldrage K, Robbins EC, et al. Faecal immunochemical tests (FIT) versus colonoscopy for surveillance after screening and polypectomy: a diagnostic accuracy and cost-effectiveness study. Gut 2019;68:1642–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin JS, Piper MA, Perdue LA, et al. Screening for Colorectal Cancer: Updated Evidence Report and Systematic Review for the US Preventive Services Task Force. Jama 2016;315:2576–94. [DOI] [PubMed] [Google Scholar]
- 43.Chiu HM, Lee YC, Tu CH, et al. Association between early stage colon neoplasms and false-negative results from the fecal immunochemical test. Clin Gastroenterol Hepatol 2013;11:832–8 e1-2. [DOI] [PubMed] [Google Scholar]
- 44.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;370:1287–97. [DOI] [PubMed] [Google Scholar]
- 45.Li M, Wu P, Yang Z, et al. miR-193a-5p promotes pancreatic cancer cell metastasis through SRSF6-mediated alternative splicing of OGDHL and ECM1. Am J Cancer Res 2020;10:38–59. [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng G, Zhang G, Zhao Y, et al. Screening of miRNAs as Prognostic Biomarkers for Colon Adenocarcinoma and Biological Function Analysis of Their Target Genes. Front Oncol 2021;11:560136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camps C, Buffa FM, Colella S, et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 2008;14:1340–8. [DOI] [PubMed] [Google Scholar]
- 48.Gee HE, Camps C, Buffa FM, et al. hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer 2010;116:2148–58. [DOI] [PubMed] [Google Scholar]
- 49.Ho AS, Huang X, Cao H, et al. Circulating miR-210 as a Novel Hypoxia Marker in Pancreatic Cancer. Transl Oncol 2010;3:109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hong L, Yang J, Han Y, et al. High expression of miR-210 predicts poor survival in patients with breast cancer: a meta-analysis. Gene 2012;507:135–8. [DOI] [PubMed] [Google Scholar]
- 51.Zhao A, Li G, Péoc’h M, et al. Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol 2013;94:115–20. [DOI] [PubMed] [Google Scholar]
- 52.Wang W, Qu A, Liu W, et al. Circulating miR-210 as a diagnostic and prognostic biomarker for colorectal cancer. Eur J Cancer Care (Engl) 2017;26. [DOI] [PubMed] [Google Scholar]
- 53.Świtlik W, Karbownik MS, Suwalski M, et al. miR-30a-5p together with miR-210-3p as a promising biomarker for non-small cell lung cancer: A preliminary study. Cancer Biomark 2018;21:479–488. [DOI] [PubMed] [Google Scholar]
- 54.Pasculli B, Barbano R, Rendina M, et al. Hsa-miR-210-3p expression in breast cancer and its putative association with worse outcome in patients treated with Docetaxel. Sci Rep 2019;9:14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mosakhani N, Sarhadi VK, Borze I, et al. MicroRNA profiling differentiates colorectal cancer according to KRAS status. Genes Chromosomes Cancer 2012;51:1–9. [DOI] [PubMed] [Google Scholar]
- 56.Mosakhani N, Pazzaglia L, Benassi MS, et al. MicroRNA expression profiles in metastatic and non-metastatic giant cell tumor of bone. Histol Histopathol 2013;28:671–8. [DOI] [PubMed] [Google Scholar]
- 57.Li W, Chang J, Tong D, et al. Differential microRNA expression profiling in primary tumors and matched liver metastasis of patients with colorectal cancer. Oncotarget 2017;8:35783–35791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muti P, Donzelli S, Sacconi A, et al. MiRNA-513a-5p inhibits progesterone receptor expression and constitutes a risk factor for breast cancer: the hOrmone and Diet in the ETiology of breast cancer prospective study. Carcinogenesis 2018;39:98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li A, Yu J, Kim H, et al. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res 2013;19:3600–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y, Zhao H, Gao X, et al. Identification of a three-miRNA signature as a blood-borne diagnostic marker for early diagnosis of lung adenocarcinoma. Oncotarget 2016;7:26070–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


