Figure 5. Comparison of age-associated changes in the transcriptome of NMR, human, and mouse skin.
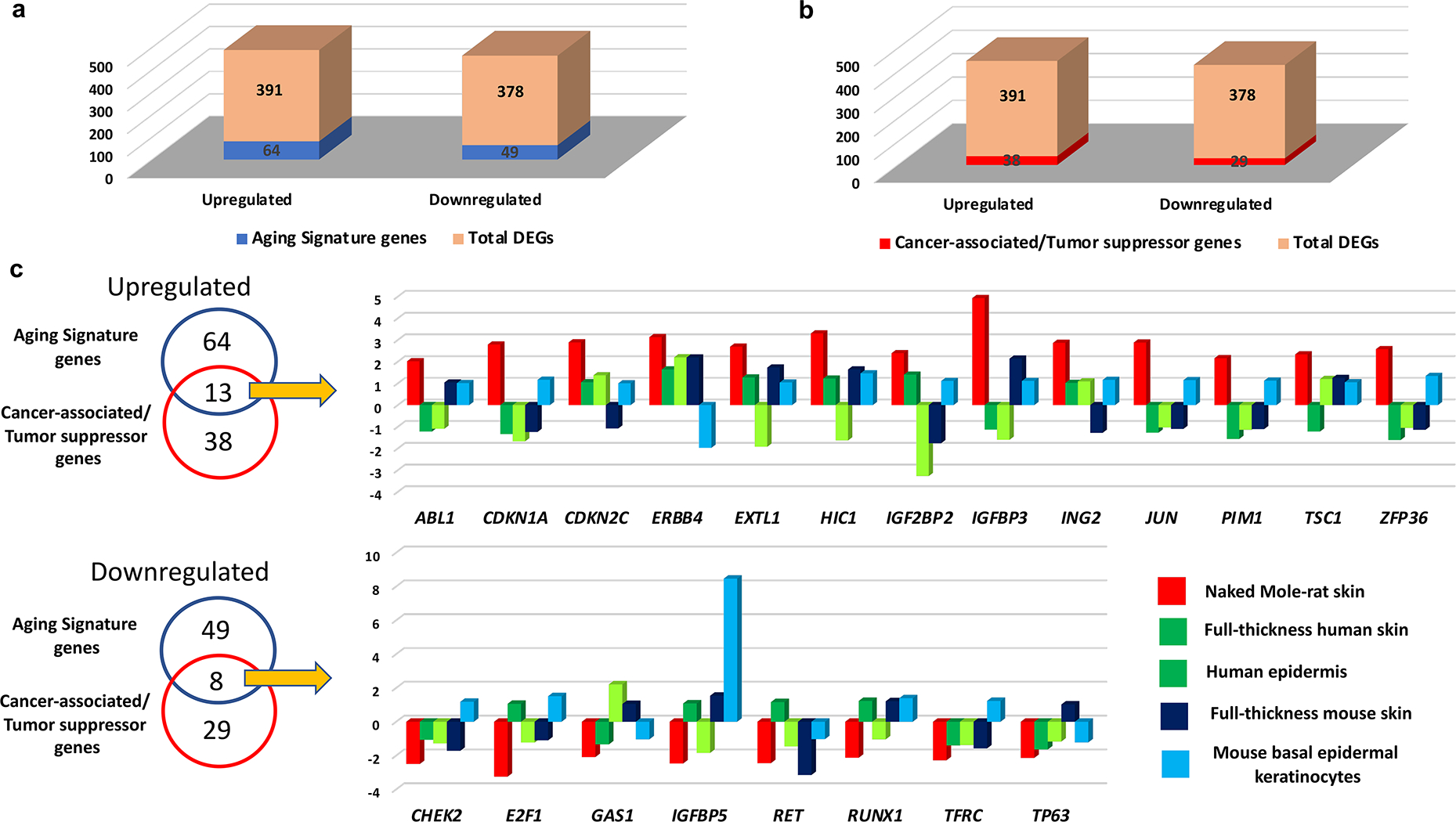
(a) The number of differentially expressed genes common between the NMR skin aging transcriptome and the Human Ageing Genome Resource (HAGR) genes (here denoted as aging signature genes) relative to the total number of differentially expressed genes; (b) The number of differentially expressed genes common between the NMR skin aging transcriptome and the Cancer Gene Census (CGC)/Tumor Suppressor Genes (TSG) datasets (here denoted as cancer signature genes) relative to the total number of differentially expressed genes; (c) A Venn diagram shows the differentially expressed genes in the NMR skin aging transcriptome that are shared with the human HAGR and CGC/TSG databases. Comparative analyses of gene expression (old versus young) in the skin of three species: NMR (total skin), human (total skin, epidermis) and mouse (total skin, FACS-sorted basal epidermal keratinocytes). The differences in gene expression are shown as the FPKM fold-change.
