Figure 6. Fibroblast Subclustering.
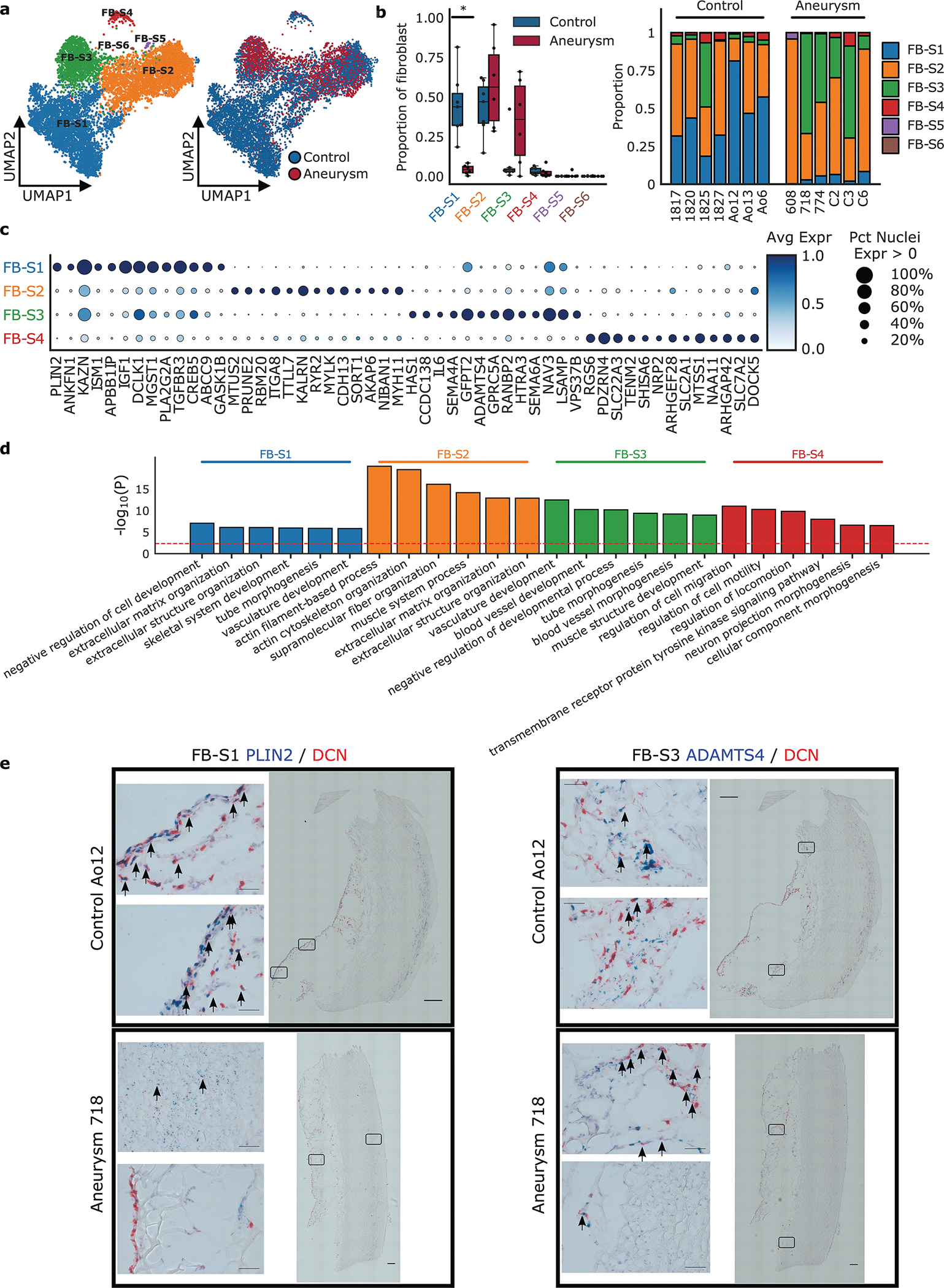
a. Nuclei clustered as fibroblast were selected to identify clusters enriched for marker genes of other global cell types. Results of subcluster analysis showing six fibroblast sub-clusters. b. The relative proportion of each subcluster between control and aneurysmal aortic tissue, and the relative proportion of each subcluster by sample. Statistically credible shifts in proportions as tested using scCODA (see Methods) are denoted with a *. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range. c. Selected marker genes that define each subcluster. d. Gene ontology pathway enrichment based on marker genes for each fibroblast sub-cluster. The red line indicates FDR < 0.05. Avg Expr, Average log-normalized expression scaled to the maximum expression in any sub-cluster; Pct Nuclei Expr > 0, Percent of nuclei in a given sub-cluster that express the gene at non-zero levels. e. RNA labeling of aortic tissue using RNAscope in situ hybridization. Control and aneurysm aortic tissue section labeled with DCN (red, global fibroblast marker), PLIN2 (blue, FB-S1 marker) and ADAMTS4 (blue, FB-S3 marker). Identified FB-S1 and FB-S3 populations appear blue and are indicated by arrows. Images show lack of FB-S1 in the aneurysm tissue, presence of FB-S3 in the aneurysm tissue, both in the adventitial layer. 40x single images = 200 um20x tiled images = 1000 um
