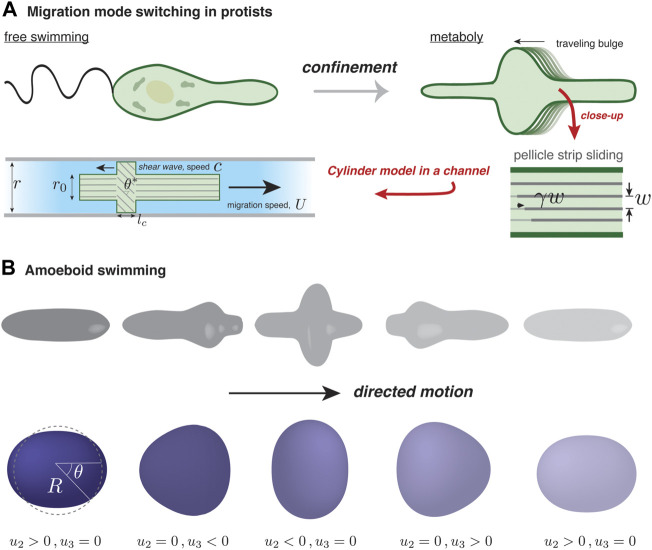
FIGURE 2.
Self-deformation-based migration. (A) Depending on confinement, protists from the genus Euglena can swim using a flagellum or inch forward using surface waves, a process known as metaboly. The rearward traveling surface bulges characterizing metaboly are generated by microtubule activity in the parallel strips forming the pellicle, an organismic envelope. The strips are assumed to maintain a fixed separation, w, and internal activity generates a localized traveling pulse of relative strip sliding, γ Noselli et al. (2019). In the simple version of the model by Noselli et al. (2019), the confined organism is modeled as a uniform cylinder of radius r 0, except over a region where γ is non-zero. There, and over a length l c , relative sliding induces tilting with respect to the channel axis and bulging of the surface to become in contact with the channel walls; the tilt angle is related to r 0 and the channel radius by cos θ* = r 0/r. In the frame of the moving cylinder, the contact zone moves rearward with speed c; the cell speed U depends solely on c and θ*. (B) Illustration of amoeboid swimming in a viscous fluid (not shown). Actively generated forces at the cell surface are assumed to give rise to cyclic cell shape changes. In order to respect Purcell’s scallop theorem Purcell (1977), directed motion requires that a forward-played movie of a “stroke” cycle looks different than its backward-played version. A minimal reduction of amoeboid swimming is a superposition of two deformation modes, one with nematic symmetry (l = 2) and another with polar symmetry (l = 3). The surface displacement away from the spherical state, u(θ, t), is projected onto the two modes, with amplitude u 2(t) and u 3(t).