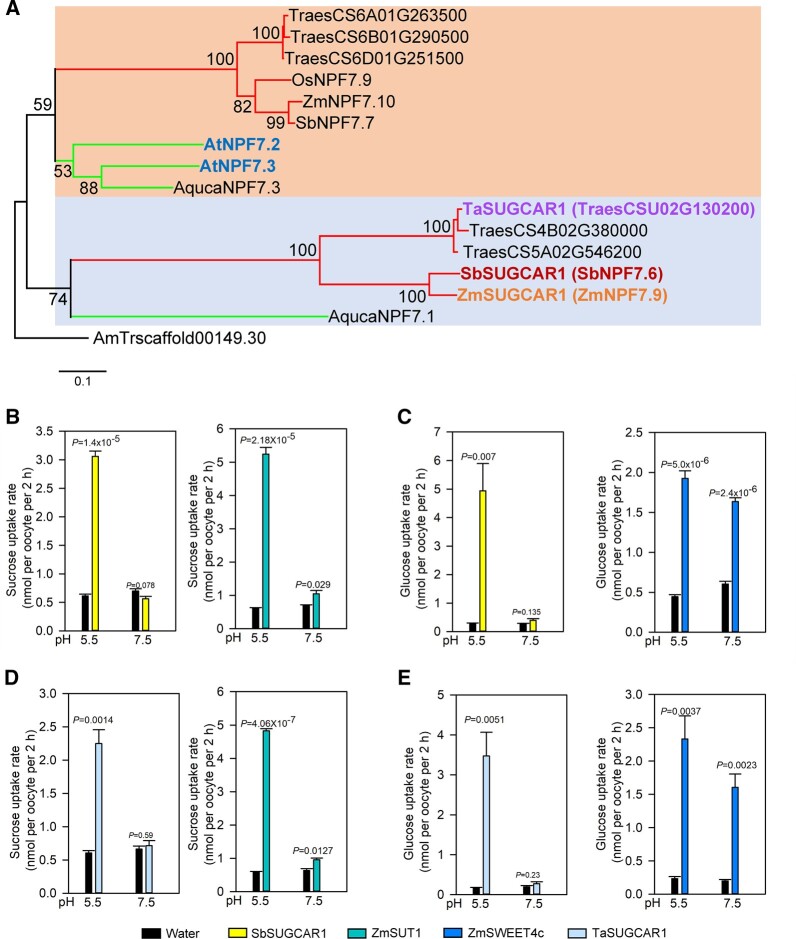
Figure 4.
Phylogenetic analysis of SUGCAR1 homologous proteins and sugar uptake by SbSUGCAR1 and TaSUGCAR1 in Xenopus oocytes. A, A simplified phylogenetic tree of SUGCAR1 homologous proteins from different plant species. The protein sequences of A. trichopod was used as outgroup. Two large, color-shaded blocks indicated major clades in this superfamily. The upper and lower branches represented monocots and dicots in each color-shaded clade, respectively. ZmSUGCAR1 (ZmNPF7.9), SbSUGCAR1 (SbNPF7.6), TaSUGCAR1, AtNPF7.3 and AtNPF7.2 were highligted in bold fonts, respectively. All bootstraps (1,000 replicates) above 50 were listed on the branches. The scale bar represented branch length. Species abbreviations were as follows: Ta or Traes, T. aestivum; Os, O. sativa; Zm, Z. mays; Sb, S. bicolor; At, A. thaliana; and Aquca, A. coerulea. Accession numbers of NPF proteins were the same as in Léran et al. (2014). B–E, Sugar uptake by SbSUGCAR1 and TaSUGCAR1 in oocytes. Oocytes injected with water were used as the negative control, and oocytes injected with ZmSUT1 or ZmSWEET4c were used as positive controls. P < 0.05 indicated significant differences.