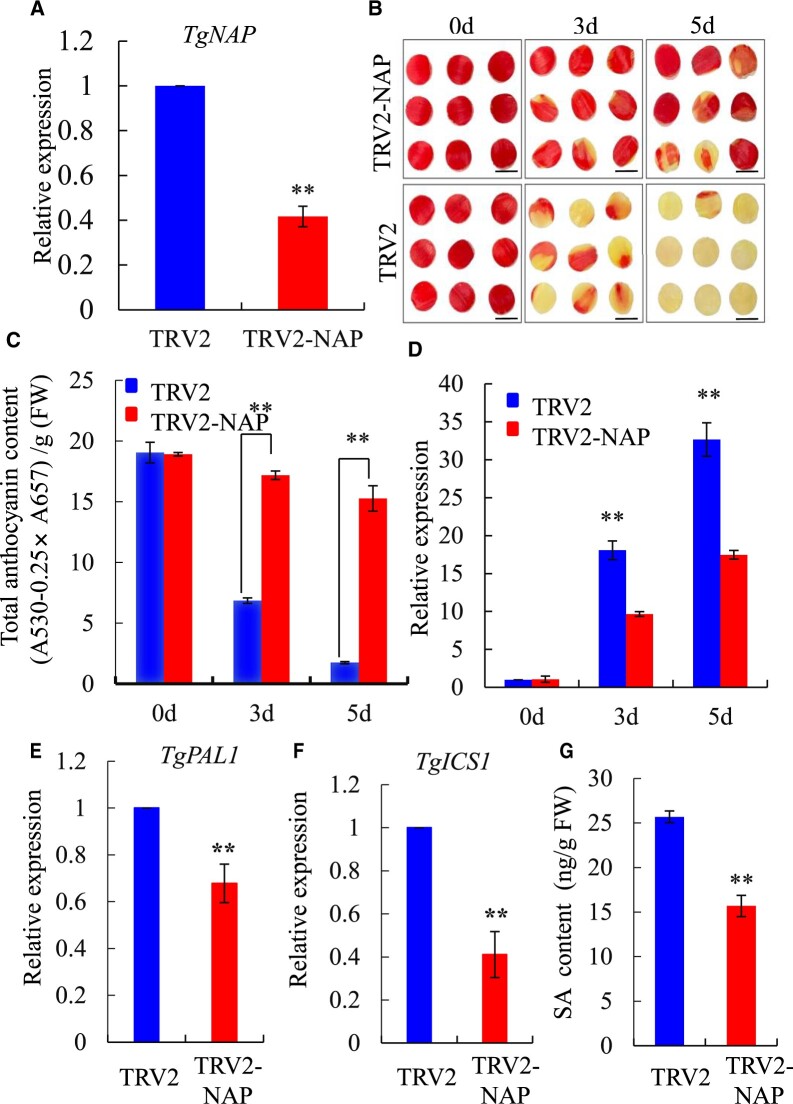
Figure 3.
Silencing of TgNAP delayed senescence in tulip petal discs. A, Expression levels of TgNAP in the TRV2 control and TgNAP-silenced petals at Day 3 by RT-qPCR analysis. The expression levels of the samples at the indicated time point were normalized to a TgUBQ10-like gene. Relative gene expression was calculated using the 2–ΔΔCt method. B, Phenotypes of tulip petal discs detached from TRV2 control and TRV2-TgNAP line under ambient conditions. Scale bar, 1 cm. The samples in each boxed area were photographed at the same time and are part of a single image. C, Total anthocyanin levels of TRV control and TgNAP-silenced petals. D, Expression levels of TgSAG6 of TRV2 control and TgNAP-silenced petals. E and F, Expression levels of TgPAL1 (E) and TgICS1 (F) of TRV2 control and TgNAP-silenced petals at Day 3. The expression levels of the samples at the indicated time point were normalized to a TgUBQ10-like gene. Relative gene expression was calculated using the 2–ΔΔCt method. G, SA content in TRV2 control and TgNAP-silenced petals at Day 5. Error bars indicate SE (n = 3). Asterisks indicate significant difference between the TgNAP-silenced petals and the TRV control based on the LSD values (**P < 0.01).