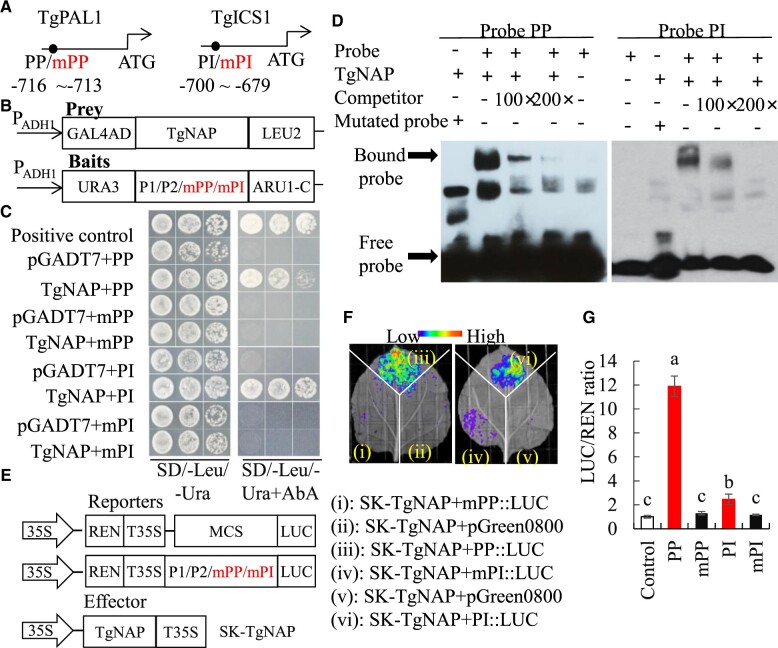
Figure 5.
Direct binding of TgNAP to promoters of TgPAL1 and TgICS1. A, Schematic diagram of the TgPAL1 and TgICS1 promoter. The black circles indicate the position of the partial promoter fragments within CACG motifs. PP: promoter of TgPAL1; PI: promoter of TgICS1. The numbers shown below indicated the position of specific binding of TgNAP in the promoters ahead of ATG. B, The prey and bait constructs used for Y1H assay. mPP and mPI is a mutated version of PP and PI, in which the CACG was replaced with AAAG. PADH1, promoter of alcohol dehydrogenase1. GAL4AD, galactose-specific transcription enhancing factor 4 activation domain. C, Growth of yeast cells co-transformed with various prey plus bait combinations on selective medium (SD/-Ura/-Leu) with (right) or without (left) AbA. AbA, Aureobasidin A. Positive control: p53-AbAi+pGAD-p53; Negative control: bait+pGADT7. D, EMSA analysis of specific binding of TgNAP to the promoter of TgPAL1 and TgICS1. The purified His-TgNAP protein and biotin-labeled probe of designed fragments containing CACG motif or mutated AAAG motif were used. Competitor was unlabeled probe at 100- and 200-fold. +: presence; −: absence. E, The schematic of the TgNAP effector and pTgICS1::LUC, pTgPAL1::LUC reporters. T35S, the terminator of CaMV 35S, respectively. MCS, multiple cloning sites. LUC, firefly luciferase. REN, Renilla luciferase. F, Live image of transcriptional activation of the TgICS1 and TgPAL1 promoters by TgNAP in N. benthamiana leaves using Dual-LUC system. G, Quantitative analysis of dual-LUC transient expression assays of the promoter activity in N. benthamiana protoplasts. LUC/REN ratio of SK-TgNAP + pGreen0800 empty vector was used as the negative control and was considered as 1 for data normalization. Error bars indicate SE (n = 3). Different letters denote significant differences at P < 0.05 analyzed by Tukey’s test.