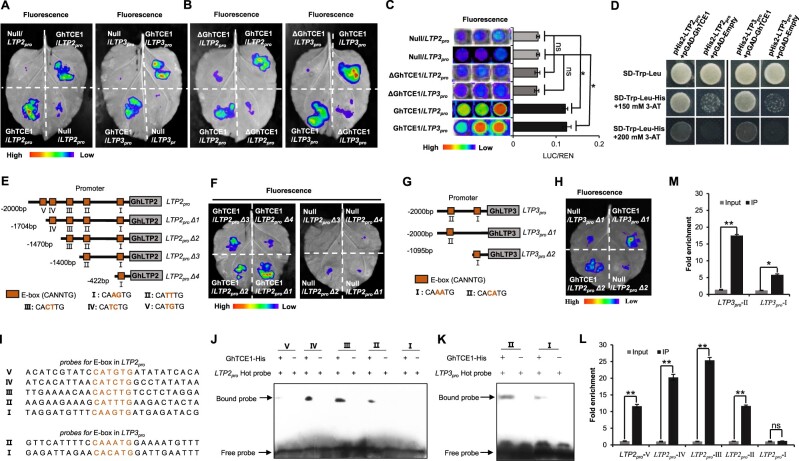
Figure 4.
GhTCE1 binds to the promoters of GhLTP2 and GhLTP3 to activate their expression. A and B, LUC signals detected in tobacco leaves. Promoters of GhLTP2 and GhLTP3 (LTP2pro and LTP3pro) were separately fused upstream of LUC in the vector pGreenII 0800-LUC and designated LPT2pro:LUC and LPT3pro:LUC. As an internal control, 35Spro:REN in vector pGreenII 0800-LUC was used. GhTCE1 and ΔGhTCE1 were fused downstream of the 35S promoter in pGreenII 62-SK. The pGreenII 62-SK empty vector was used as negative control and named Null. For each tobacco leaf, half was used to express pGreenII 0800-LUC:LTPpro+pGreenII 62-SK:GhTCE1+35Spro:REN (named GhTCE1/LTPpro) and half was used to transiently express pGreenII 0800-LUC:LTPpro+pGreenII 62-SK empty vector:REN (named Null/LTPpro) following Agrobacterium-mediated transfection, ΔGhTCE1 in the same way. C, Activated expression of LTP2pro and LTP3pro by GhTCE1 in cotton callus protoplasts using the LUC expression system. Relative activation level represents the ratio of fluorescence intensity of LUC/REN with GhTCE1 to LUC/REN without GhTCE1. D, Y1H assays of GhTCE1 binding to LTP2pro and LTP3pro. The full-length coding sequence of GhTCE1 was fused to the pGADT7 vector, and LTP2pro and LTP3pro were cloned into the pHis2 vector. Interaction was determined on selective medium lacking Trp+Leu+His in the presence of 3-AT. E, A series of truncations (LTP2proΔ1 to LTP2proΔ4) with E-box deletions were used in the LUC activation system vector. F, GhTCE1 activates the expression of truncated LTP2pro by detected LUC signals on tobacco leaves. The truncated LTP2pro constructs were fused to the vector pGreenII 0800-LUC, and combined with pGreenII 62-SK:GhTCE1+35Spro:REN as a positive control. The pGreenII 62-SK empty vector was used as a negative control. They are labeled the same way as in (A) and (B). G, GhTCE1 activates the expression of truncated LTP3pro (LTP3proΔ1 andLTP3proΔ2) by LUC signals detected on tobacco leaves. H, GhTCE1 activates the expression of truncated LTP3pro (LTP3proΔ1 and LTP3proΔ2) by LUC signals detected on tobacco leaves. The process same to (F). I, The probe sequences of LTP2pro and LTP3pro are noted and the E-boxes are shown in red. J and K, EMSA showing the specificity of in vitro binding between GhTCE1 and the E-box elements of LTP2pro (J) and LTP3pro (K). The WT probes were subjected to biotin-labeling. No GhTCE1-His tag protein lines as control. The arrows indicate the up-shifted binds. L and M, Validation of the binding of GhTCE1 to the promoters E-box elements of LTP2pro (L) and LTP3pro (M) by ChIP-qPCR. Error bars in (C), (L), and (M) represent ± standard errors of three biological replicates (10 samples each in [C], three samples each in [L] and [M]). Statistical significance is indicated by *P < 0.05, **P < 0.01, Supplemental Data Set S2.