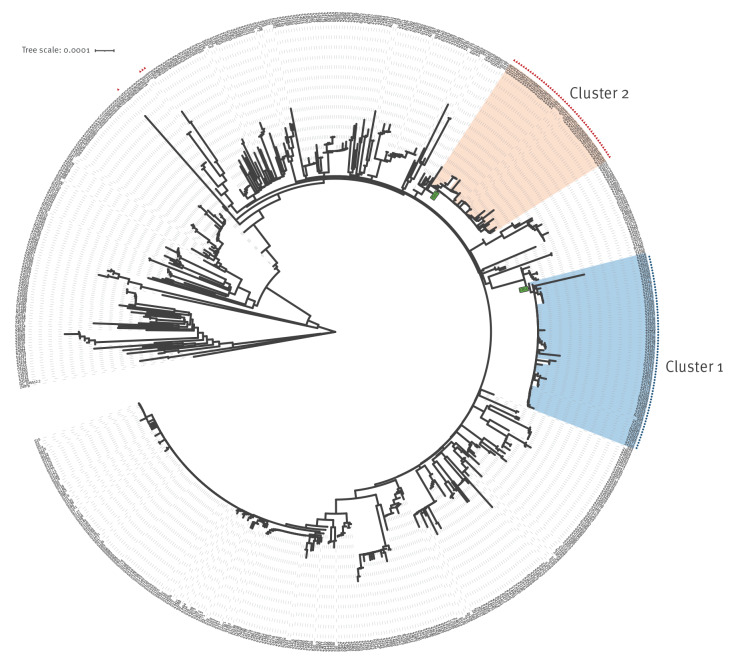
Figure 1.
Phylogenetic analysis of the extended dataset of sequences of SARS-CoV-2 registered cases, Düsseldorf and Solingen, Germany, 15 June–01 August 2021 (n = 699 sequences)a
SARS-CoV-2: severe acute respiratory coronavirus 2.
a The phylogenetic tree, generated with Geneious (see Methods) and visualised with iTol [19], shows 699 sequences. Nine sequences were found to exhibit low-quality alignments in the multiple sequence alignment of the 708 input sequences by manual inspection and were removed from the alignment before construction of the tree.
Cluster 1 and Cluster 2 are highlighted (areas of the tree shaded in blue and red, respectively). The nodes serving as the two clusters’ root nodes, I361 and I584, are displayed as little boxes with green background. The presence of T14064C and C18744T, mutations used in the process of defining Cluster 1 and Cluster 2, is indicated by blue circles and red triangles, respectively.