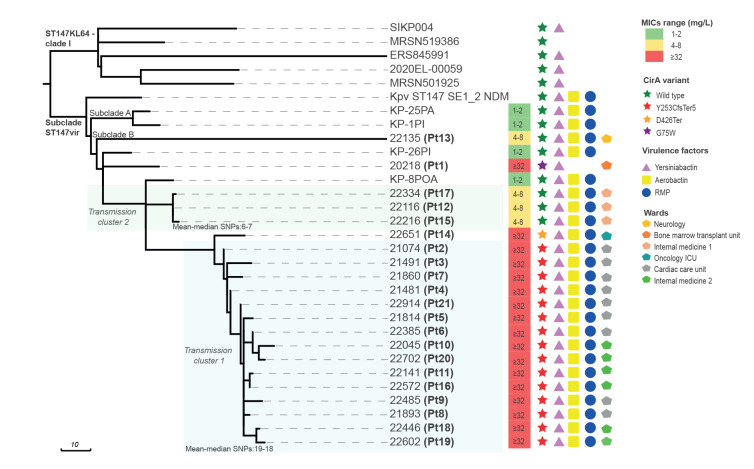
Figure 3.
Phylogenetic analysis of cefiderocol-resistant NDM-1-producing ST147 Klebsiella pneumoniae from tertiary care hospital outbreak in Florence, Italy, August 2021–June 2022 (n=21)
ICU: intensive care unit; MIC: minimum inhibitory concentration; RMP: regulators of mucoid phenotype.
KP-1PI, KP-25PA, KP-26PI, KP-8POA were included as comparators belonging to the ST-147-vir clone circulating in Tuscany [9]. ERS845991 (from Hungary), MRSN519386 (from Middle East region), SIKP004 (from Thailand), MRSN501925 (from Germany), Kpv_ST147_SE1_2_NDM (from United Kingdom) and 2020EL-00059 (from United States) strains were included in the analysis as outliner representants of ST147 K. pneumoniae. The short-term bacterial evolution was generated using Gubbins v. 3.2.1 and RaxML tree builder. Branch lengths were reported in substitutions (point mutations) per genome. The complete genome of K. pneumoniae KP-1PI (Accession number CP071027), that was the first NDM-Kp ST147 isolated from a Tuscan region outbreak in 2018, was used as reference FDC susceptible strains for all in silico analyses [9].