FIGURE 5.
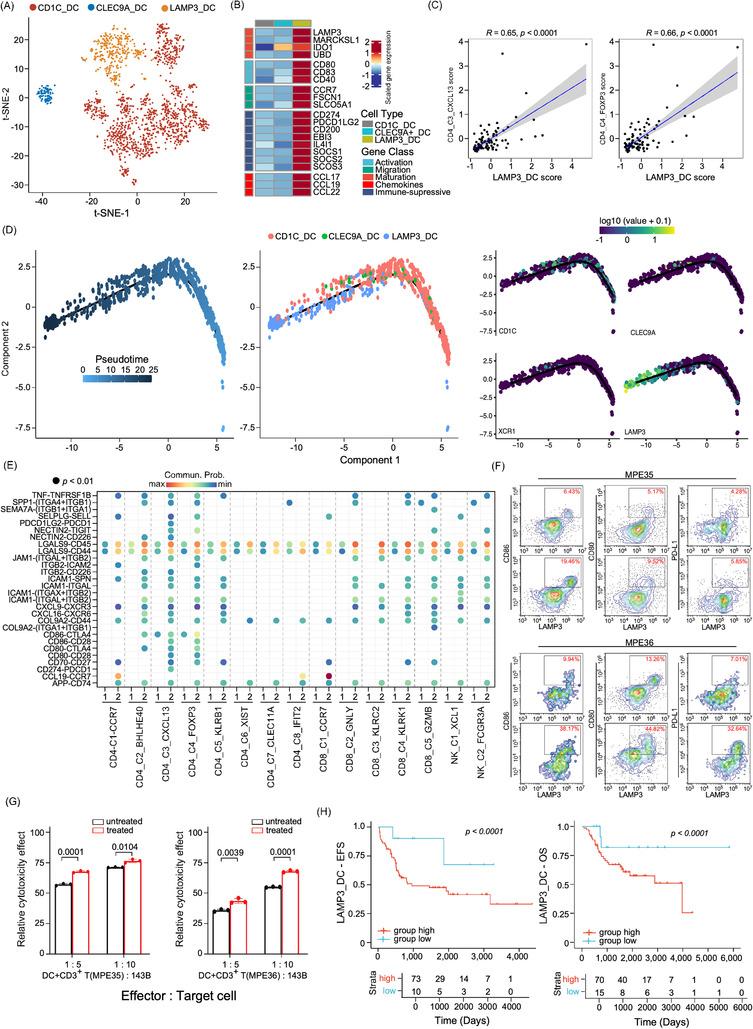
Characterization of dendritic cell (DC) sub‐clusters in osteosarcoma patients. (A) The t‐SNE plot showed the three DC sub‐clusters in osteosarcoma patients as coloured. (B) The heatmap displayed the normalized mean expression of genes associated with activation, migration, maturation, chemokines and immuno‐suppressive activities in the three DC clusters. (C) Scatter plot presented the correlation between LAMP3_DC and CD4_C4_FOXP3 Treg or CD4_C3_CXCL13 gene signature scores in TARGET‐osteosarcoma patient cohort (n = 85). (D) Trajectory analysis plots of DC cell functional state transition, coloured by pseudotime (left panel), cell sub‐cluster (middle panel) or the expression levels of biomarker genes, including CD1C, CLEC9A, XCR1 and LAMP3 (right panel), respectively. (E) Dotplot displayed the comparison of the ligand–receptor interaction of LAMP3_DC and with T/NK sub‐clusters in MPE and primary tumour tissues. 1 indicated primary tumour (PT) and 2 indicated MPE samples. (F) Flow cytometry plot exhibited LAMP3, CD80, CD86 and PD‐L1 levels in DCs treated with tumour alloantigens in two MPE samples (MPE35 and MPE36). The upper panel shows the untreated control group, and the lower panel shows DCs loaded with tumour alloantigens. (G) The cytotoxicity test showed a significant difference between the control and treated groups. A comparison was performed using Student's t‐test. (H) The Kaplan–Meier plot demonstrated event‐free survival (EFS) and overall survival (OS) of TARGET‐osteosarcoma patients (n = 85) categorized by the LAMP3_DC gene signature score. The x‐axis represents time (days) and the y‐axis represents survival probability. Comparisons between curves were performed using log‐rank tests.
