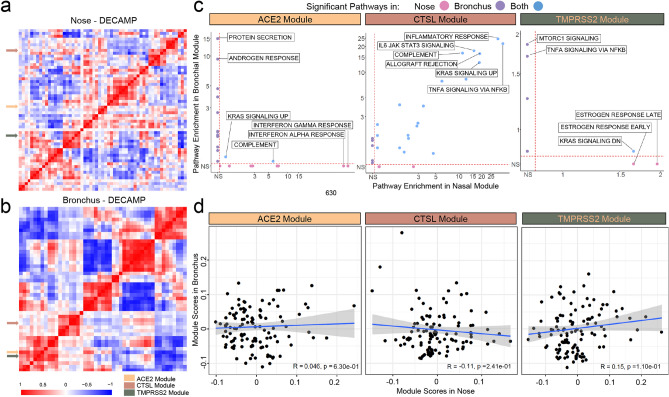
Figure 2.
Consensus nasal and bronchial VE gene modules are associated with different biological pathways. (a–b) WGCNA was used to identify consensus co-expression modules in nasal (a) and bronchial (b) samples from the DECAMP cohort (n = 58 and 48 modules, respectively). A heatmap of the correlation of module eigengenes computed from each module demonstrates that ACE2 and TMPRSS2 modules were more highly correlated with each other in the bronchus than in the nose. The CTSL module was not correlated with ACE2 or TMPRSS2 modules in either the nose or bronchus. (c) Scatterplots comparing the overrepresentation of MSigDB Hallmark pathway gene sets in each VE module in the nose and bronchus. The -log10(FDR q) values denoting the degree of overrepresentation of each gene set in each module in the nose and bronchus are shown on the x- and y-axes, respectively. The overrepresentation of gene sets in the ACE2 and TMPRSS2 modules is largely discordant between the nose and bronchus, whereas several immune-related pathways are overrepresented in the CTSL module in both sites. (d) VE module eigengenes were not significantly correlated (Pearson p > 0.05) between paired nasal (x-axis) and bronchial (y-axis) samples in DECAMP (n = 114). The blue line is the line of best fit and the gray shading represents the 95% confidence level interval for predictions from the linear model.