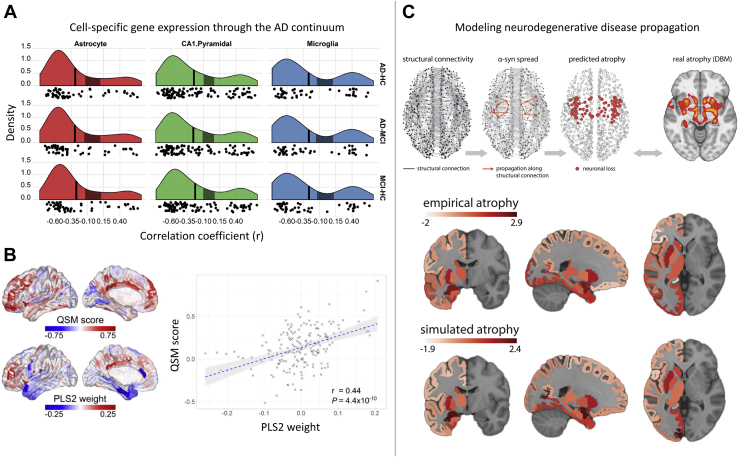
Figure 4.
Gene expression is related to neurodegeneration and neurodegenerative disease spread. (A) Cell-specific marker association with cortical thinning in AD. Correlation distributions quantifying the relationship between cell-specific gene expression profiles for CA1 pyramidal, microglia, and astrocyte cell types and cortical thickness reductions in AD (patients with AD vs. HCs; AD vs. MCI; MCI vs. HC). Vertical axis denotes estimated probability density for the correlation coefficients. Vertical black line denotes the average correlation coefficient across all genes; shaded gray box indicates the 95% limits of the empirical null distribution. (B) Gene expression is associated with cortical iron decomposition, quantified using QSM, in Parkinson’s disease. Cortical maps represent QSM scores and regional linearly weighted sum of gene expression scores defined by PLS2. (C) Schematic representation of pathology spread across the brain: misfolded α-synuclein propagates via structural connections; simulated neuronal loss (atrophy) is compared against empirical atrophy, estimated from patients with Parkinson’s disease. Simulated and empirical atrophy patterns show a high level of spatial correspondence (Spearman correlation, r = 0.63, p = 2 × 10−5). Panel (A) adapted with permission from (81). Panel (B) adapted with permission from (121). Panel (C) adapted with permission from (132). α-syn, α-synuclein; AD, Alzheimer’s disease; CA, cornu ammonis; DBM, deformation-based morphometry; HC, healthy control; MCI, mild cognitive impairment; PLS, partial least squares; QSM, quantitative susceptibility mapping.