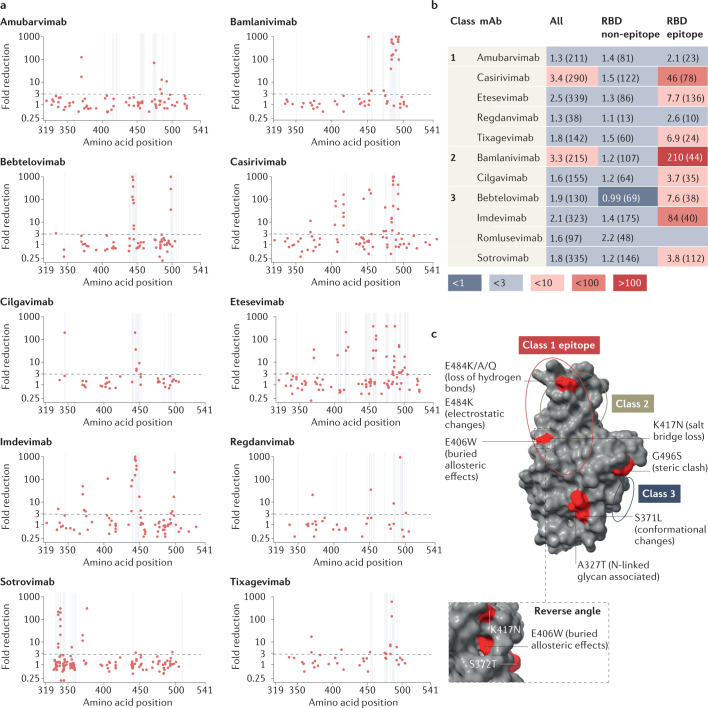
Fig. 3. Assessment of monoclonal antibody resistance by mutations of interest occurring at epitope and non-epitope sites.
a, Geometric mean fold reduction in neutralization (mFRN) data for each monoclonal antibody (mAb) against viral constructs containing single mutations in the spike receptor binding domain (RBD) (positions 319–541). Epitope positions (see Supplementary Data File 3) indicated by vertical grey lines. Dashed line shows the mFRN = 3 threshold. Alternative substitutions at the same amino acid position are shown as separate points at the same x coordinate, for example E484K and E484Q are shown as two different points at the 484 position on the x axis. Most dots above the threshold coincide with the grey lines, indicating that mutations conferring resistance to neutralization tend to occur at epitope positions. However, some non-epitope mutations also confer resistance as shown by the dots above the threshold but not on grey lines. b, Pooled mFRN comparisons between epitope and non-epitope mutations. Values shown are mFRN for either all amino acid positions, all amino acid positions in the epitope of the mAb, or all amino acid positions in the RBD but not in the epitope of the mAb. The higher mFRN values of epitope positions indicate the increased contribution of mutations in the mAb epitope to the resistance of neutralization. Number in brackets indicates the number of assays contributing to each geometric mean value. Epitope unknown for romlusevimab. Colours depict the strength of resistance: dark red, strong (mFRN > 100); red, moderate (mFRN = 10–100); light red, mild (mFRN = 3–10); light grey, no resistance (mFRN = 1–3); dark grey, increased sensitivity (mFRN < 1). c, Isolated Omicron spike RBD structure [PDB:7TGW]98. Epitope regions for class 1, 2 and 3 mAbs are circled. Epitope and non-epitope mutations exemplifying mechanisms of mAb evasion are labelled: S371L, conformational changes and N-linked glycosylation43; A372T, N-linked glycosylation50; E406W, conformational changes in the epitope46; K417N, abolished salt bridges between mAb and RBD41; E484K/E484Q/E484A, loss of hydrogen bonds with mAb56; E484K, changes to electrostatic interactions54; G496S, steric clash99. Single mutant mFRN data across the full spike protein are shown in Supplementary Fig. 6a and pooled mFRN comparisons between epitope, non-epitope mutations, epitope proximal and RBD positions in Supplementary Fig. 6b.