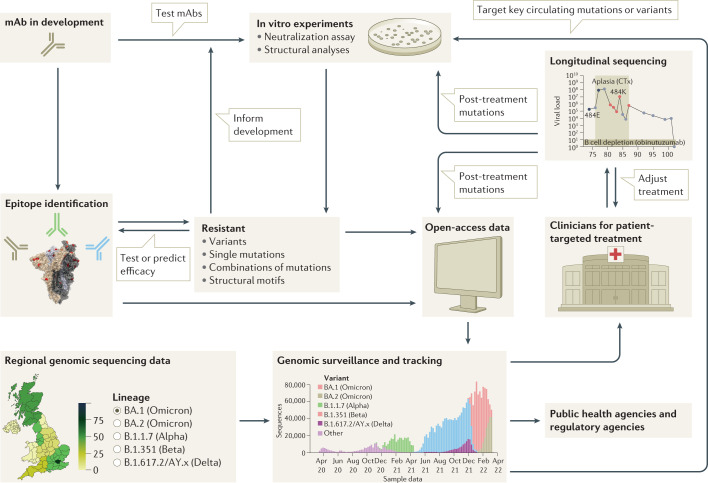
Fig. 4. Framework for the rational design of viral genomic surveillance for the development of efficient monoclonal antibody therapies.
A successful framework will integrate knowledge of the dynamics of monoclonal antibody (mAb) resistance by SARS-CoV-2 variants, including the central role played by epitope mutations, epistatic effects and evolutionary dynamics. mAb development must be supported by epitope identification, in vitro studies and the monitoring of resistance mutations. mAbs in early development may be ruled out if they target an epitope that contains mutations shown to cause resistance in vitro. Alternatively, low frequency of, or evolutionary barriers to, mutations in a given region may focus the development of mAbs that target that region. Resistance monitoring is a multifaceted process, spanning individual and combinatorial mutational effects in vitro, as well as genomic surveillance of circulating strains and longitudinal clinical studies that track genetic changes during treatment. Combining these different approaches will allow the development of mAbs highly effective against currently circulating strains, and robust to resistance by future variants.