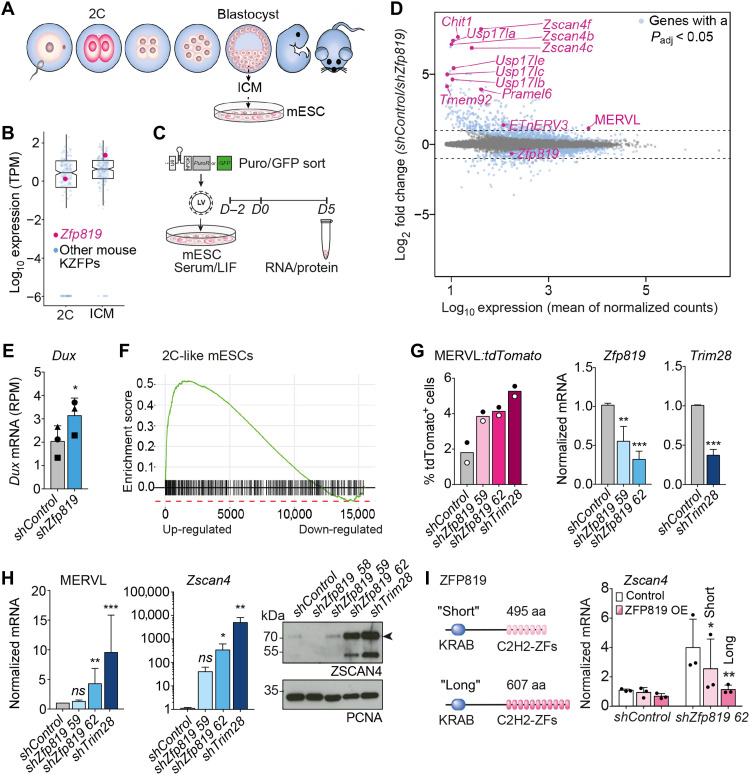
Fig. 1. mESCs transition to a 2C-like state upon loss of ZFP819.
(A) mESCs are used to model reprogramming to a state with similarities to 2C embryos. ICM, inner cell mass; 2C, 2-cell. (B) Transcript levels of 327 KZFPs, including Zfp819, in a 2C embryo and the blastocyst. KZFPs, KRAB zinc finger proteins; TPM, transcripts per million. (C) Experimental setup: shRNAs are added on day −2. LIF, leukemia inhibitory factor. (D) Differentially expressed genes and transposable element (TE) families upon ZFP819 depletion, compared to control. Events beyond the dashed lines represent log2 fold changes >1 or < −1. Highlighted in blue are genes with a P-adjusted value < 0.05. 2C genes are labeled in pink. (E) RNA-seq reads mapping to Dux in control and ZFP819-depleted cells. P = 0.0451 (two-tailed paired Student’s t test). RPM, reads per million. (F) Gene set enrichment analysis of 2C-like genes (3). P-adjusted value = 0.0002. (G) Left: MERVL-tdTOMATO–positive cells, following ZFP819 depletion (mean of two independent experiments). Right: RT-qPCR analysis of Zfp819 and Trim28 transcripts, following their shRNA depletion (mean ± SD of three independent experiments). P values: 0.0090 (shZfp819 59), 0.0005 (shZfp819 62), and 0.0005 (shTrim28) (two-tailed paired Student’s t tests). (H) RT-qPCR validation of 2C genes up-regulated (left) and Western blot of ZSCAN4 protein (right). One representative experiment of three is shown. RT-qPCR data show mean ± SD of three independent experiments. P values for MERVL: 0.0093 (shZfp819 62) and 0.0008 (shTrim28). P values for Zscan4: 0.0415 (shZfp819 62) and 0.0022 (shTrim28) (two-tailed unpaired Student’s t tests). ns, not significant. (I) Domains of ZFP819 Short and Long isoforms (left). RT-qPCR detection of Zscan4 (right): Rescue experiments were performed with non–HA-tagged constructs. Data are mean ± SD of three independent experiments with replicate values shown. P values: short isoform = 0.0357; long isoform = 0.0011 (two-tailed paired Student’s t tests). aa, amino acids.