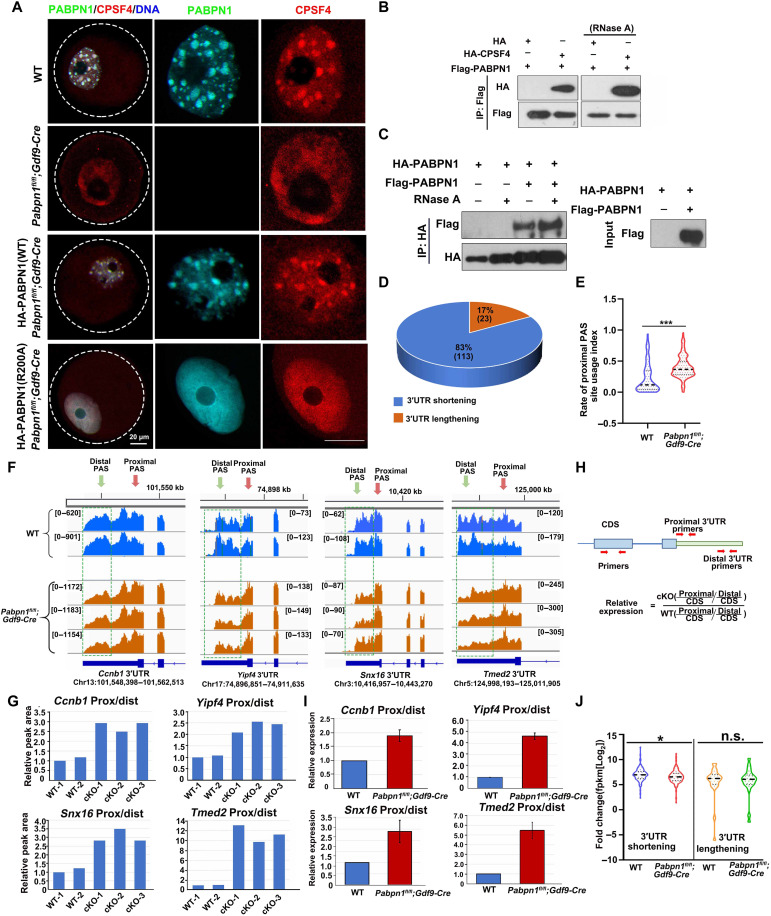
Fig. 9. Transcript 3′UTR selection in Pabpn1-knockout oocytes.
(A) Immunofluorescence results showing the distribution of PABPN1 and CPSF4 in WT and Pabpn1fl/fl;Gdf9-Cre mouse oocytes. Scale bar, 20 μm. (B and C) Co-IP experiment showing the interaction of PABPN1 with CPSF4 and PABPN1 (C). (D) Number of transcripts with 3′UTR shortening or lengthening in Pabpn1fl/fl;Gdf9-Cre mice oocytes, as compared to those in control oocytes. (E) Comparison between the proximal PUI calculated in WT and Pabpn1fl/fl;Gdf9-Cre mouse oocytes. The analysis included 136 transcripts that showed a significant shift in the PAS usage. (F) Examples of transcripts showing significant 3′UTR shortening, manifested by prominent decrease peaks near the distal CS relative to the proximal ones in Pabpn1-deletion oocytes, as compared to those in WT samples. Transcripts with green boxes represent the distal CS levels. Green arrow, distal PAS; red arrow, proximal PAS. (G) The ratios of the proximal to distal peak area are shown in (F), normalized to the average of the two controls. (H) The primer design pattern diagram detects the proportion of long and short isomers in the transcript. Separately designed primer sequences to detect the CDS, proximal, and distal isoforms of a specific transcript. The proportions of the proximal and distal isomers of the same transcript were normalized to the CDS. (I) To confirm the results of the 3′UTR selection, the same samples as in (F) were subjected to qRT-PCR. (J) RNA sequencing results showing the relative levels of transcripts that displayed 3′UTR shortening or lengthening in WT and Pabpn1fl/fl;Gdf9-Cre mice oocytes. Error bars represent the SEM. *P < 0.05 by as assessed using the two-tailed Student’s t test.