Figure 5.
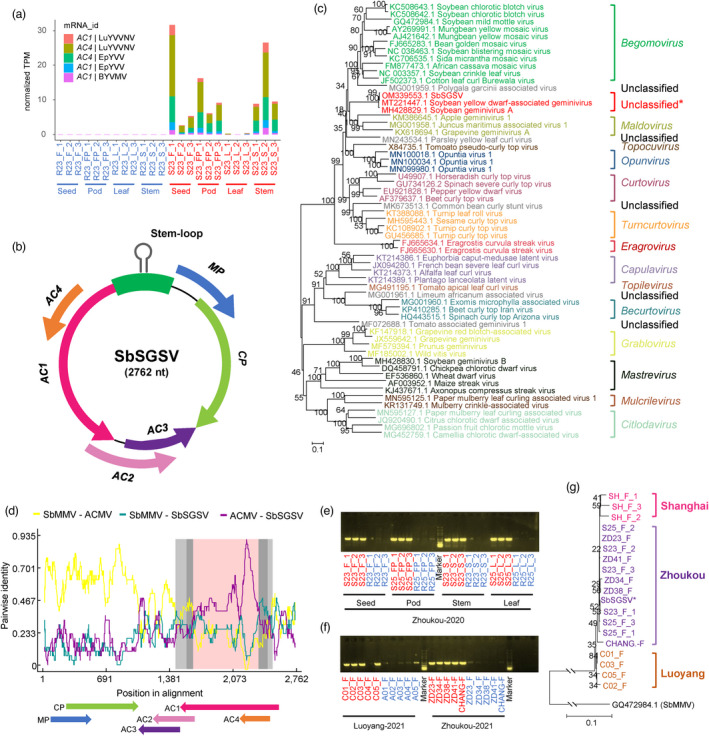
Molecular characterization of a geminivirus associated with the incidence of soybean SGS. (a) AC1 and AC4 gene of Begomovirus were particularly higher expressed in SGS diseased tissues. Red and blue labels represent diseased and healthy samples, respectively. (b) Schematic representation of SbSGSV genomic organization. Open reading frames organization on the genome sequence are denoted with different colours. (c) Phylogenetic relationships of SbSGSV and representative geminiviruses assigned to the 14 established geminivirus genera based on the nucleotide sequence of the full‐length genome using the neighbour‐joining method. The statistical significance of the branches was determined with a bootstrap of 1000 replicates. (d) Recombination analysis of SbSGSV. The green line indicates the pairwise identity between the SbMMV (Soybean mild mottle virus, GQ472984.1) and SbSGSV, the brown line indicates the pairwise identity between the ACMV (African cassava mosaic virus, FM877473.1) and SbSGSV. The grey region represents the location of predicted breakpoints (99% (grey) and 95% (darkgrey) breakpoint confidence interval, the pink region represents the tract of sequence with a recombinant origin. (e) PCR gel band showing the presence or absence of SbSGSV in the healthy (blue) and SGS diseased (red) tissues. (f) PCR gel band showing the presence or absence of SbSGSV in the healthy (blue) and SGS diseased (red) seeds in YD22 in Luoyang, and in diverse soybean cultivars (ZD23, ZD34, ZD38, ZD41 and a descendant cultivar CHANG18008‐2‐6) in Zhoukou. For (e and f), DNA fragments of 734 bp were amplified using primers Ss_f12‐Ss_r12. (g) Phylogenetic relationships of SbSGSV and amplicon sequences of geminivirus associated with SGS in diverse soybean cultivars across three different locations (Zhoukou, Luoyang and Shanghai). Soybean mild mottle virus (SbMMV, GQ472984.1) was used as outgroup. The neighbour‐joining tree was constructed using the 734 bp sequences amplified by primers Ss_f12‐Ss_r12. The statistical significance of the branches was determined with a bootstrap of 1000 replicates.
