Figure 2.
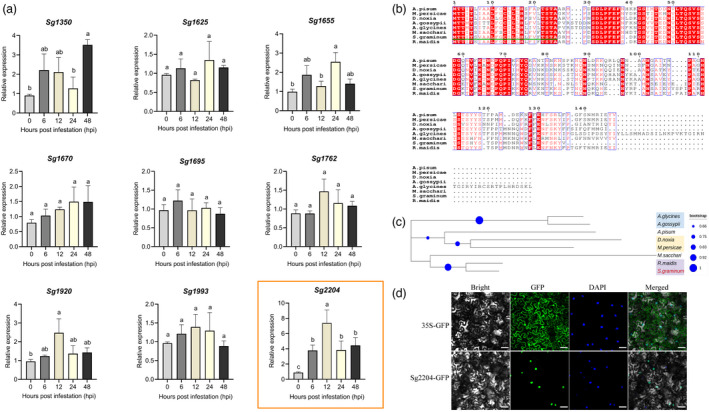
Characterization of Sg2204 in Schizaphis graminum. (a) RT‐qPCR results of the relative expression levels of nine candidate effectors of S. graminum after infestation on wheat plants at different time points. Data are presented as the mean ± SE (n = 3). Different lowercase letters above the bars indicate significant differences among groups (one‐way ANOVA followed by Duncan's multiple range tests, P < 0.05). (b) Multiple sequence alignment of the Sg2204 protein and orthologs from other aphid species. The deduced amino acid sequences from seven aphid species, including Acyrthosiphon pisum (NP_001156070.1), Myzus persicae (XP_022163343.1), Diuraphis noxia (XP_015374910.1), Aphis gossypii (XP_027851062.1), Aphis glycines (KAE9525059.1), Melanaphis sacchari (XP_025207447.1) and Rhopalosiphum maidis (XP_026819965.1). The sequence of Sg2204 was submitted to NCBI GenBank under the accession number OM676656. Red shades indicate identical amino acids. Alignment was created using Clustal Omega. (c) Phylogenetic tree constructed by comparing the amino acid sequences of Sg2204 and the orthologs from other aphid species. A phylogenetic tree was constructed by the neighbour‐joining method using MEGA7.0. Bootstrap values calculated as a percentage for 1000 replications are shown at the nodes. The amino acid sequence with green underline indicates the signal peptide of the Sg2204 protein. (d) Subcellular localization of Sg2204. GFP (empty vector, pCAMBIA1300) and effector Sg2204 were transiently overexpressed in N. benthamiana leaves by Agrobacterium infiltration. The images were taken 3 days after agroinfiltration using confocal microscopy. Bar = 20 μm.
