Figure 1.
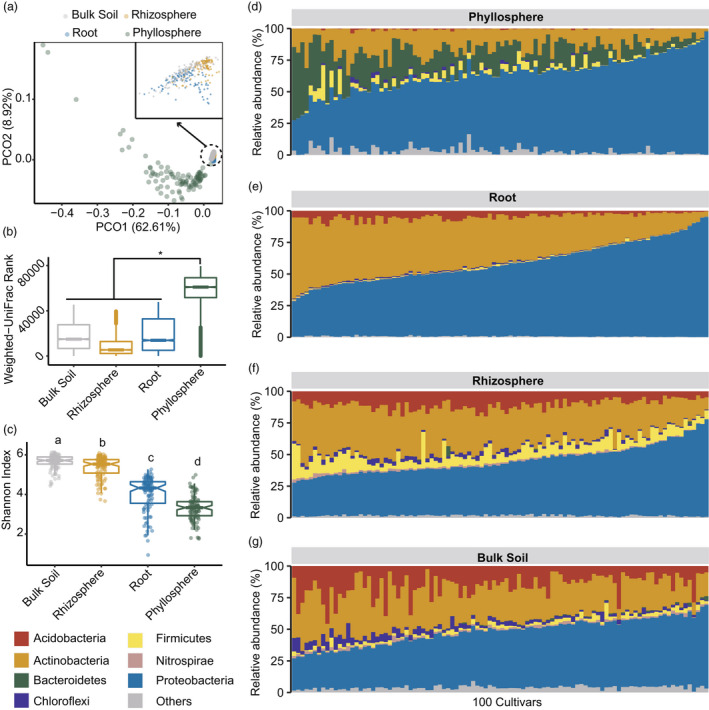
Diversity and composition of microbial communities in 100 tea (Camellia sinensis) cultivars. (a) Principal coordination analysis (PCoA) for the microbiomes in bulk soil, rhizosphere soil, root and phyllosphere compartments (n = 100 for each compartment). Eigenvectors of the top two principal coordinates (PCOs) were used to visualize the microbial β‐diversity. The samples of bulk soil, rhizosphere and root were zoomed‐in for clarity purposes. (b) Comparison of microbial weighted UniFrac dissimilarity between phyllosphere and the other three compartments by the analysis of similarity (ANOSIM, R = 0.91, P = 0.001). (c) Alpha diversity (Shannon index) of the microbiomes in different compartments. The letters above the boxes indicated Tukey's test after one‐way ANOVA (n = 100 for each compartment). (d–g) Microbial composition of 100 tea cultivars in phyllosphere (d), root endosphere (e), rhizosphere (f) and bulk soil (g). The seven most abundant microbial phyla for each compartment were shown and other phyla were aggregated into ‘others’.
