FIG 2.
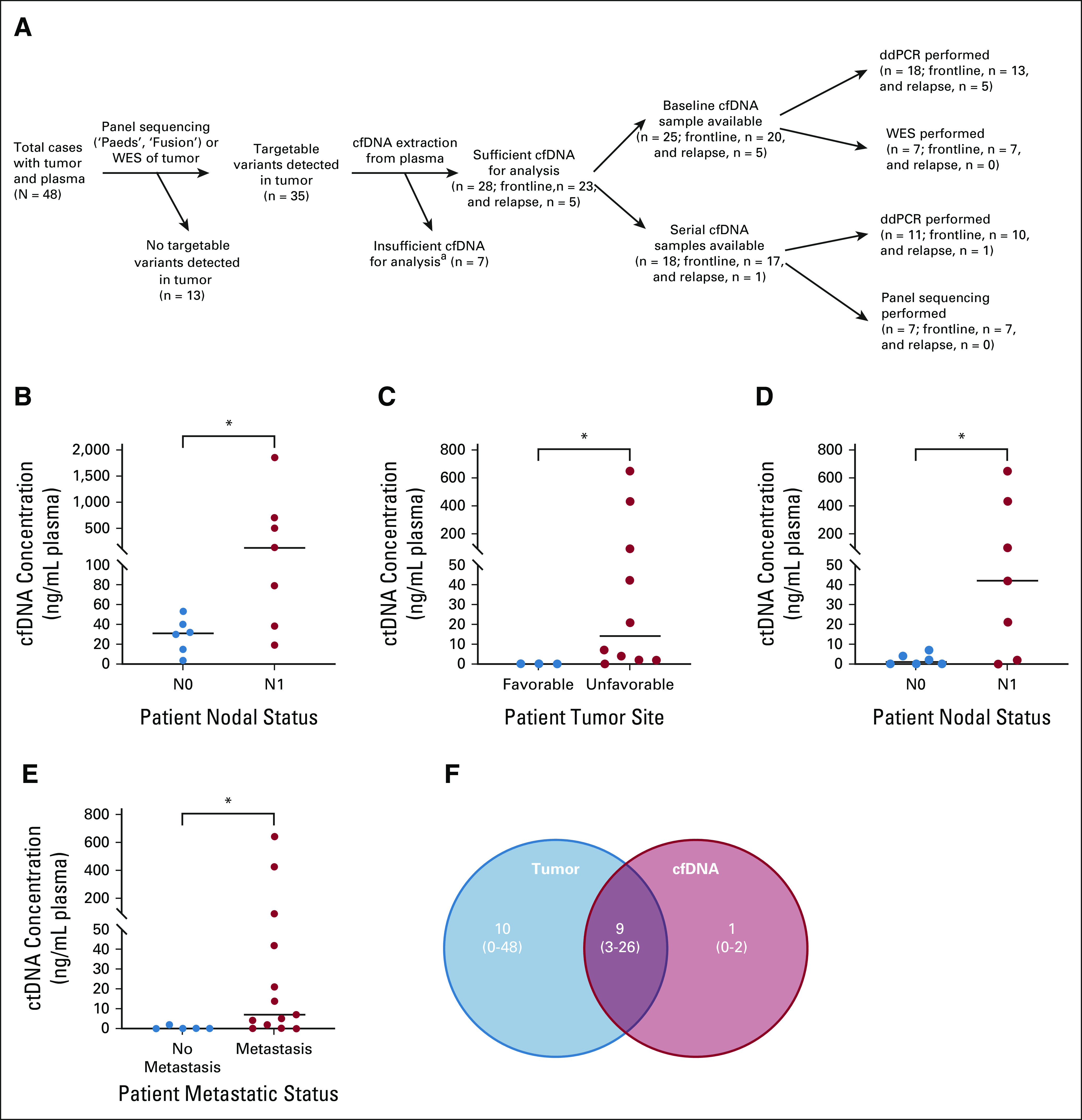
Baseline cfDNA analysis. (A) Overview of samples collected (n = 48 tumor and plasma) and successfully analyzed (n = 28 cfDNA). aInsufficient cfDNA yield is defined as < 1 ng for ddPCR and < 10 ng for WES/panel sequencing. (B) Frontline patients with nodal spread (N1, n = 7) had significantly higher baseline cfDNA yields (ng/mL plasma) compared with those without it (N0, n = 6; P = .035). Baseline ctDNA yields (ng/mL plasma) were significantly higher in frontline patients with (C) tumors in an unfavorable site (n = 10) compared with those with tumors in a favorable site (n = 3; P = .0210) and (D) nodal involvement (N1, n = 7) compared with those without it (N0, n = 6; P = .043). (E) Baseline ctDNA yields (ng/mL plasma) were significantly higher in both frontline and relapsed patients with metastases at diagnosis (n = 13) compared with those without it (n = 5; P = .0201). Median cfDNA or ctDNA yields indicated by horizontal lines on graphs. (F) There was considerable overlap in the molecular profile of matched patient tumor DNA and baseline cfDNA, as illustrated by mean number (and ranges) of variants detected in each via WES. cfDNA, cell-free DNA; ctDNA, circulating tumor DNA; ddPCR, droplet digital polymerase chain reaction.
