FIG 5.
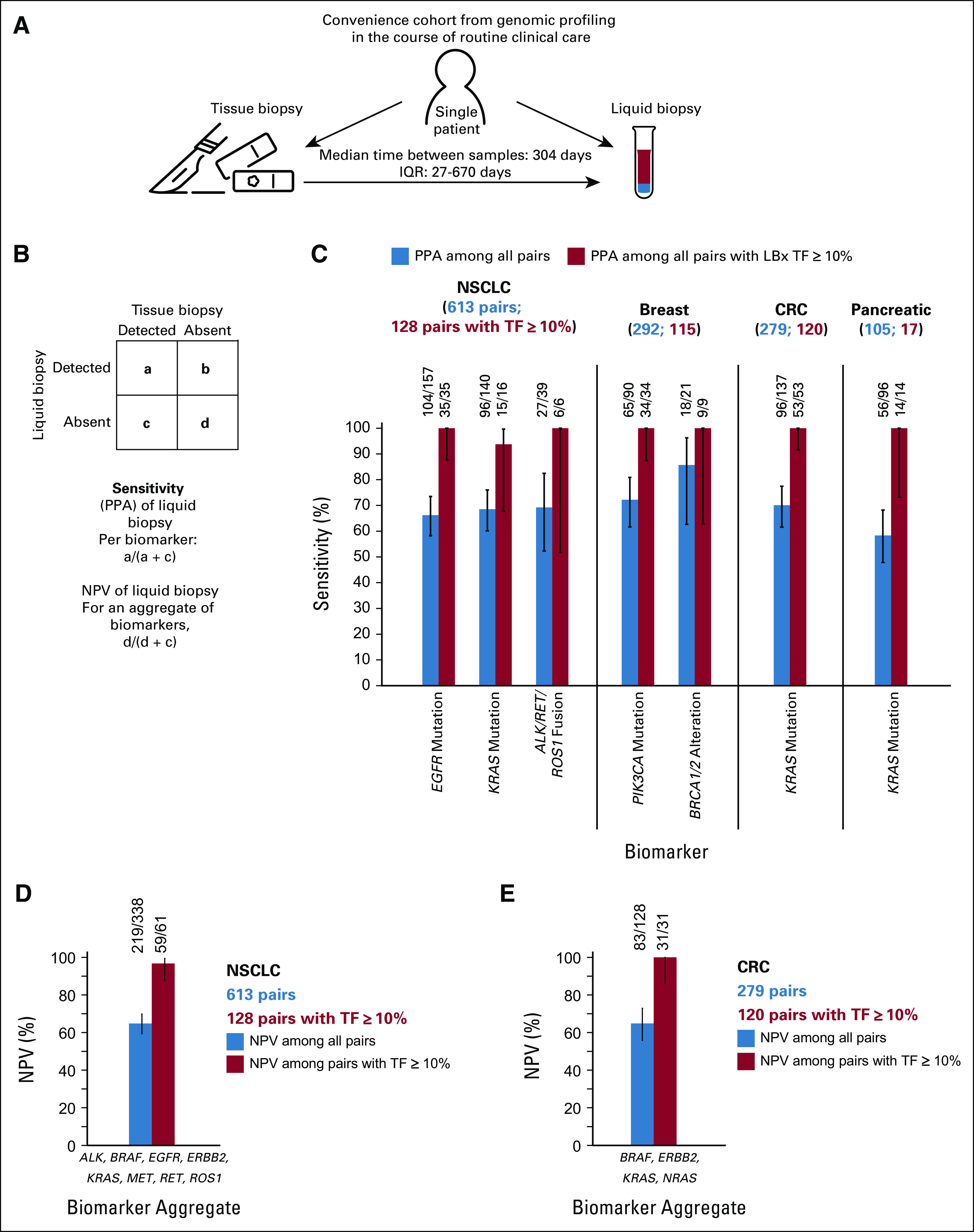
Sensitivity (PPA) and NPV of LBx profiling. Schematic of the convenience cohorts selected from the larger cohort. (A) A consecutive cohort of patients who had results from both tissue and liquid CGP was analyzed. Sensitivity of LBx (PPA) was defined for each individual biomarker with tissue as the reference standard. (B) For NPV of LBx, tissue negative for all tested biomarkers was used as the reference standard. (C) The sensitivity of LBx for detecting mutations and fusions ranged from 58% for KRAS in pancreatic cancer to 86% for BRCA1/2 alterations in breast cancer (blue bars); in the subsets of paired liquid and tissue where LBx ctDNA TF was ≥ 10%, the sensitivity of LBx was > 90% (red bars). (D) The NPV for mutations and fusions in actionable NSCLC biomarkers in eight NCCN genes (EGFR, ERBB2, KRAS, MET, ALK, BRAF, RET, and ROS1) was calculated for 613 NSCLC pairs. (E) Similarly, the NPV for mutations in actionable CRC biomarkers in four genes (BRAF, ERBB2, KRAS, and NRAS) was calculated for 279 CRC pairs. CGP, comprehensive genomic profiling; CRC, colorectal cancer; ctDNA, circulating tumor DNA; IQR, interquartile ranger; LBx, liquid biopsy; NCCN, National Comprehensive Cancer Network; NPV, negative predictive value; NSCLC, non–small-cell lung cancer; PPA, positive percent agreement; TF, tumor fraction.
