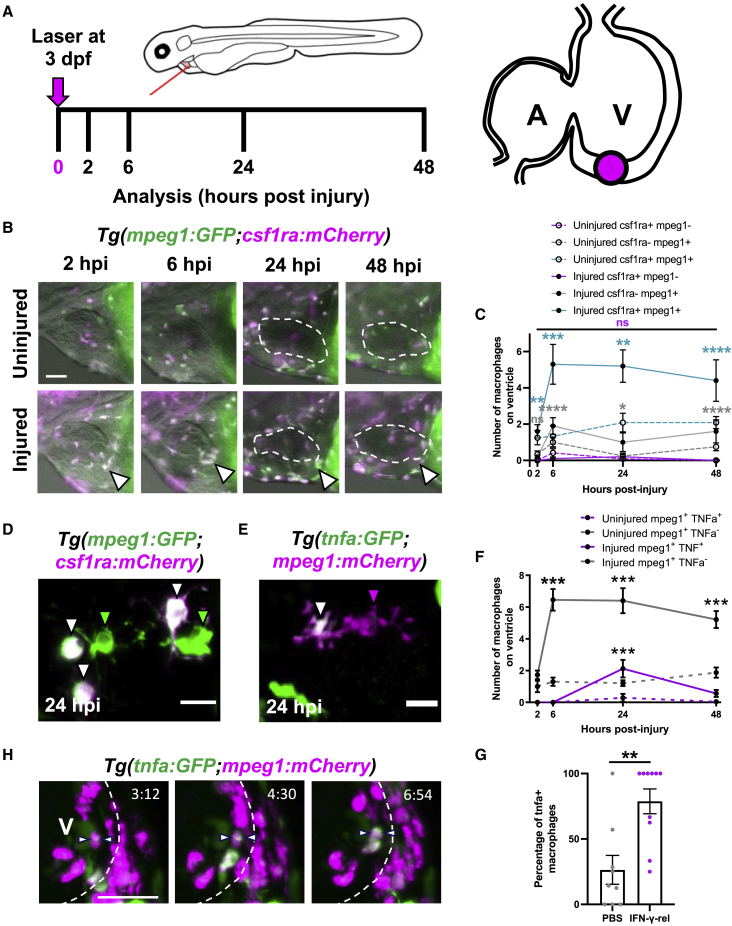
Figure 1.
Cardiac macrophages display heterogeneity and plasticity following injury
(A) Schematic illustrating the cardiac laser injury model, with imaging time points marked (left) and the injury site at ventricular apex of a 3 dpf larval heart marked (magenta circle) (right).
(B) Representative lateral-view epifluorescence images of uninjured and injured hearts at the standard time points in Tg(mpeg1:GFP;csf1ra:gal4:UAS:NfsB-mCherry) (abbreviated to mpeg1:GFP;csf1ra:mCherry in all panels) illustrating macrophage heterogeneity; white arrow, ventricular apex; dashed line, heart outline.
(C) Quantification of the number of csf1ra+mpeg1−, csf1ra-mpeg1+, and csf1ra+mpeg1+ macrophages on the ventricle in uninjured and injured larvae at standard time points, n = 10–12.
(D) Representative LSFM image of csf1ra−mpeg1+ and csf1ra+mpeg1+ macrophages of different morphologies.
(E) Representative LSFM image of tnfa+mpeg1+ and tnfa−mpeg1+ macrophages.
(F) Quantification of the number of tnfa+mpeg1+ and tnfa−mpeg1+ macrophages on the ventricle in uninjured and injured larvae at standard time points, n = 10–25.
(G) Quantification of the percentage of tnfa+ macrophages at 24 hpi following injection with IFN-γ-rel or PBS, n = 10.
(H) Time-lapse time points for injured Tg(tnfa:GFP;mpeg1:mCherry) ventricles imaged live in the larvae by heartbeat-synchronized LSFM microscopy illustrating macrophage plasticity. Timestamps indicated, dashed line, ventricle outline; arrows, macrophage converting to tnfa+. Scale bars, 50 μm in (B) and (H) and 10 μm in (D) and (E). ∗∗ p ≤ 0.01, ∗∗∗ p ≤ 0.001, ∗∗∗∗ p ≤ 0.0001 (C and F). Two-way ANOVA followed by Holm-Sidak’s post-hoc test and (G) t test. Data are represented as mean ± SEM.