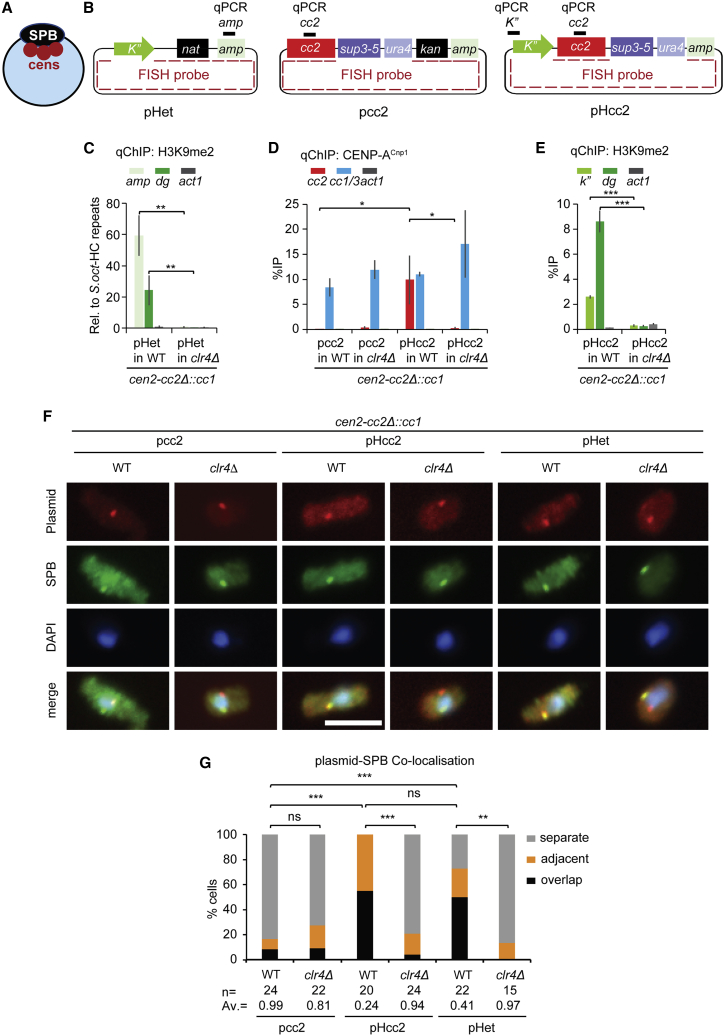
Figure 1.
Centromeric heterochromatin colocalizes with the SPB-centromere cluster
(A) Carton showing clustering of three endogenous centromeres (red circles) at the SPB (black oval) during interphase.
(B) Diagram of pHet, pcc2, and pHcc2 minichromosomes. Black bars above each plasmid map represent qChIP primer sites on ampicillin gene (amp), cc2, and K″ repeats of plasmids, respectively. Dashed red line in plasmids indicates position of FISH probe.
(C–E) qChIP analyses for H3K9me2 levels (C and E) on amp gene of pHet (C); K″ repeats of pHcc2 (E); dg repeats of centromeric HC and act1 gene; CENP-ACnp1 levels (D) on cc2, cc1/3 (indicates sequences common to cc1 and cc3), and act1 in WT and clr4Δ cells containing cc2Δ::cc1 at cen2 transformed with pHet (C), pcc2 (D), or pHcc2 (D and E). %IP levels in S. pombe were normalized to %IP of cen3 HC repeats from spiked-in S. octosporus chromatin in (C). qChIP results in (D) and (E) are reported as %IP. Data are mean ± SD (error bars) (n = 3–4 experimental replicates). ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005 (unpaired t test).
(F) Representative images of plasmid DNA FISH (red; probe as indicated in A), SPB location (green; anti-Cdc11), and DNA staining (blue, DAPI) in WT and clr4Δ cells transformed with pcc2, pHcc2, or pHet. Images were scaled relative to the maximum values of histogram. Scale bars, 5 μm.
(G) Cells were classified into three groups according to the 3D distances between plasmid and SPB (Cdc11): overlap (≤0.3 μm), adjacent (0.3–0.5 μm), or separate (0.5–3 μm).
Percentage of interphase cells (n, number analyzed from 3 independent experiments) in each category. AV, average distance; ns, no significance; ∗∗p < 0.001, ∗∗∗p < 0.0001 (Mann-Whitney U test) (see also STAR Methods and Figures S1–S3).