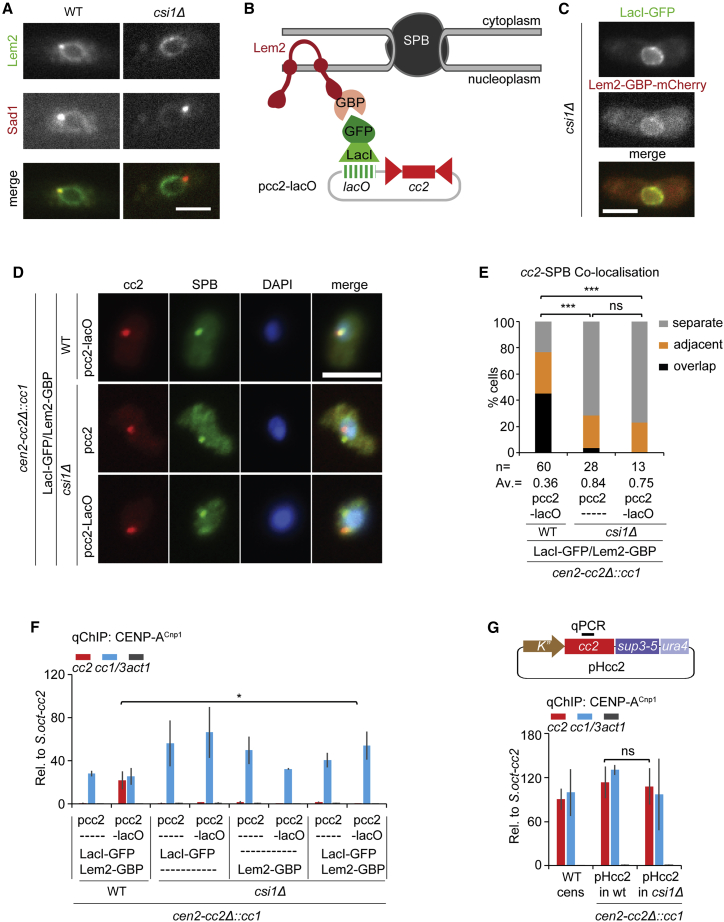
Figure 6.
Loss of Csi1 prevents CENP-ACnp1 chromatin establishment on Lem2-tethered pcc2-lacO
(A and C) Representative images of live WT and csi1Δ cells expressing Lem2-GFP and Sad1-dsRed (A) or LacI-GFP and Lem2-GBP-mCherry (C). Images were scaled as in Figure 2. Scale bars, 5 μm.
(B) Forced association of pcc2-lacO with Lem2-GBP-mCherry at NE in csi1Δ using same tethering system as in Figure 5. In csi1Δ, pcc2-lacO is expected to detach from the SPB due to loss of Lem2 from SPB.
(D) Representative images of cc2 DNA FISH (red), SPB location (green, anti-Cdc11), and DNA staining (blue, DAPI) WT or csi1Δ strains expressing both LacI-GFP and Lem2-GBP-mCherry transformed with pcc2 or pcc2-lacO. Images were scaled as in Figure 1. Scale bars, 5 μm.
(E) Percentage of interphase cells (n, number analyzed) displaying distinct degrees of cc2 DNA colocalization with SPBs (Cdc11). Cells were classified into three groups as in Figure 1. AV, average distance. ns, no significance; ∗∗∗p < 0.0001 (Mann-Whitney U test).
(F and G) qChIP analyses for CENP-ACnp1 at cc2, cc1/3, and act1 in indicated strains transform with pcc2 or pcc2-lacO (F) or pHcc2 (G). qChIP primer site on pHcc2-borne cc2 is indicated as black bar above plasmid map (G). %IP levels in S. pombe were normalized to %IP of S. octosporus central core from spiked-in chromatin. Data are mean ± SD (n = 3). ns, no significance; ∗p < 0.05 (unpaired t test) (see also Figure S1).