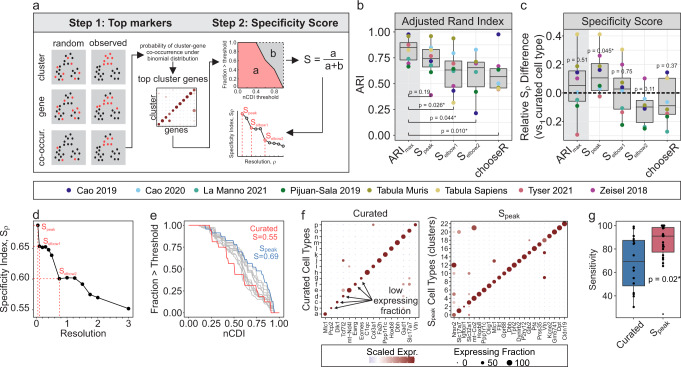
Fig. 3. Identification of optimal clustering resolution using a specificity-based criterion.
a Schematic of specificity-based resolution-selection criteria. b, c Adjusted Rand index (b) and specificity index differences (c) between ground truth (author-curated) clusters and observed Louvain clusters, using resolutions determined by different optimization criteria [specificity criteria () and chooseR58]. represents resolution at which maximal ARI was achieved, after considering all candidate resolutions (0.5–3). For b, significance compared to ARImaxdetermined by paired Wilcoxon test. For c, significance compared to zero (i.e., ground truth) was determined by one-sample Wilcoxon test. N = 8 independent scRNA-seq datasets. d–g Optimal clustering resolution for Tyser 2021 human gastrulation scRNA-seq data17. d Relationship between resolution and specificity indices, and identification of , and . e Specificity-curves. Gray curves: all candidate resolutions evaluated (0.5–3), blue curve: Speak, red curve: ground truth (curated) clusters. f Dot plots of top markers for curated (left) and (right) clusters. g Comparison of cluster sensitivity (expressing fraction) of each top marker obtained for curated and clusters. Significance determined by unpaired Wilcoxon test.