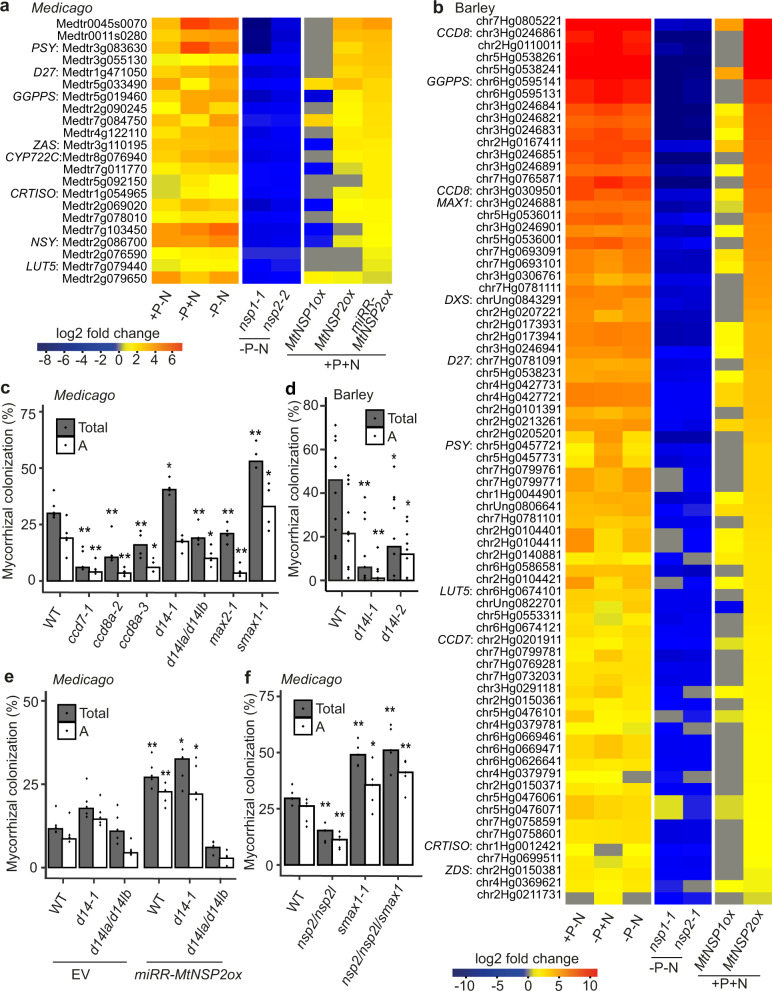
Fig. 5. NSP2 overrides P-suppression via strigolactone production and D14L/SMAX1 signaling.
Heatmaps showing N- and/or P-starvation induced genes regulated by NSP1 and NSP2 and constitutively activated by miRNA-resistant MtNSP2 overexpression in M. truncatula (a) and MtNSP2 overexpression in barley (b). Genes involved in strigolactone biosynthesis are annotated. +P–N, −P + N, and −P−N represent the expression of these starvation-induced genes in wild-type plants by comparing −N or/and −P conditions to +P + N. The nsp mutants show gene expression in nsp mutants compared to wild-type plants under nutrient depletion, while NSPox shows NSP overexpression roots compared to wild type under nutrient-replete condition. c AMF colonization at 3 weeks post-inoculation of M. truncatula mutants defective in strigolactone biosynthesis and signaling, as well as karrikin signaling. Plants were grown under low P conditions. n = 4–5 biologically independent samples. d Root-length colonization at 7 weeks post-inoculation in barley wild type and d14l mutant roots under P deficient conditions. n = 10 biologically independent plants. e AMF colonization of wild type, d14, and the d14l double mutant with or without overexpression of miRR-MtNSP2. Plants were grown under high-P (0.5 mM PO43−) conditions for 5 weeks. EV: empty vector. n = 5 biologically independent samples. f AMF colonization at 3 weeks post inoculation of M. truncatula nsp2/nsp2l, smax1, and nsp2/nsp2l/smax1 triple mutants. Plants were grown under low-P conditions. n = 5 biologically independent samples. Total: total colonization; A: arbuscules. For statistical analysis, a one-sided Wilcoxon test was performed. **p < 0.01; *0.01 < p < 0.05.