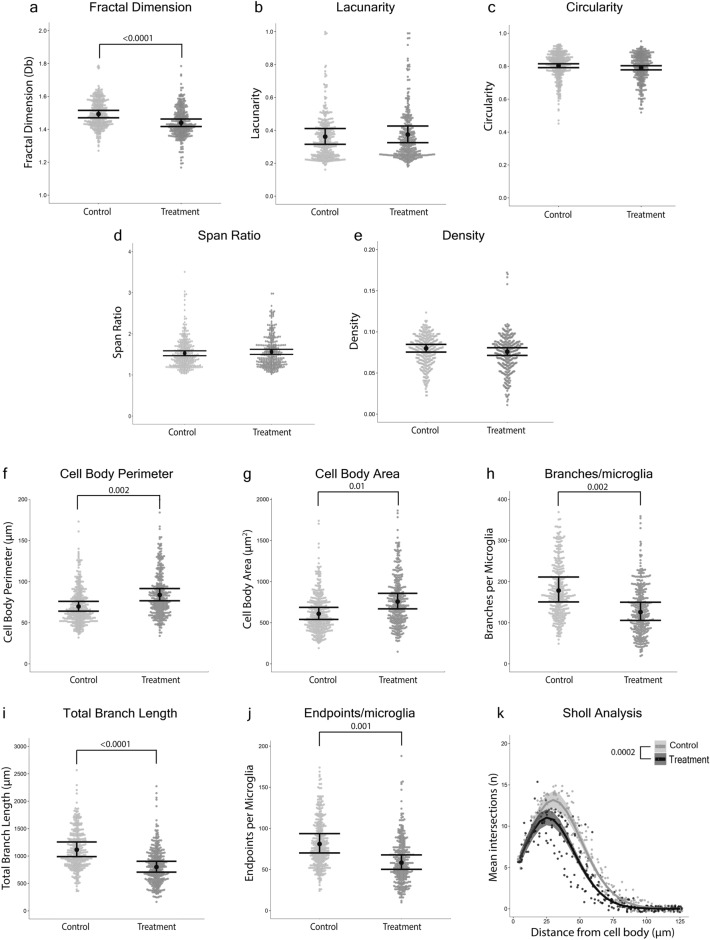
Figure 4.
Quantitative methods that analyzed microglial morphology at a single cell level were sensitive to differences between control and treatment groups. Fractal analysis detected that microglia from the treatment group had a less complex branching pattern (fractal dimension) than controls. (a) Fractal dimension (Db) of microglia; (b) lacunarity of microglia; (c) circularity of microglia; (d) span ratio of microglia; and (e) pixel density per microglia. Individual data points are presented as gray dots. Results are presented as point estimates with 95% confidence intervals, which were estimated from mixed effects models with Beta, Gaussian (fractal dimension), or Gamma (span ratio) error distributions. Individual skeletal analysis detected that microglia from the treatment group had larger cell bodies and less ramification than controls. (f) Perimeter of cell bodies; (g) area of microglia cell bodies; (h) number of branches per microglial cell; (i) total branch length per microglial cell; and (j) number of branch endpoints per microglial cell. Individual data points are presented as gray dots. Results are presented as point estimates with 95% confidence intervals, which were estimated from mixed effects models with a negative-binomial error distribution. Sholl analysis detected that microglia from the treatment group were less ramified than controls. (k) The mean number of intersections on concentric circles every 5 µm from the cell body per microglial cell. Individual data points are presented as gray dots. Results are presented as point estimates with 95% confidence intervals, which were estimated from a mixed effects model with a negative-binomial error distribution. Treatment n = 324 microglia (12 mice, 27 microglia/mouse), Control n = 345 microglia (13 mice, 27 microglia/mouse).