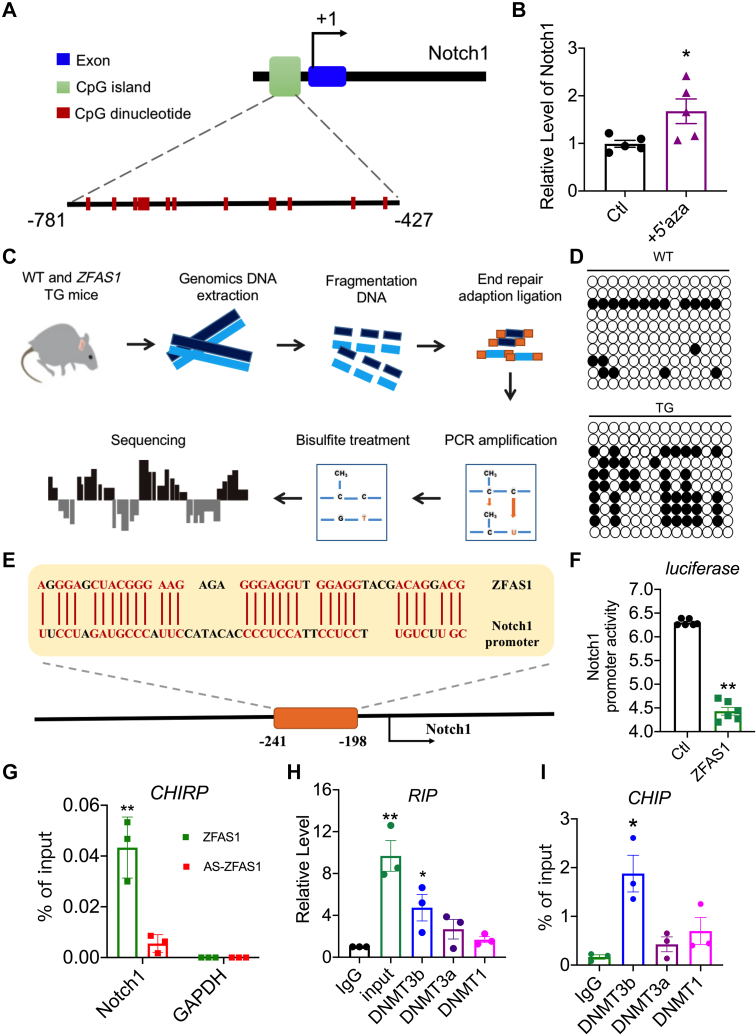
Figure 6.
ZFAS1 Promotes DNA Methylation of Notch1
(A) The Notch1 gene’s genomic architecture is depicted in a diagram. (B) The expression level of Notch1 in NMCMs treated with 5-aza-2′-deoxycytidine (5′aza). n = 5. ∗P < 0.05 vs Ctl. (C) Schematic diagram of the bisulfite sequencing. (D) Bisulfite PCR analysis of the Notch1 promoter methylation level in WT and TG mice. (E) The binding region between ZFAS1 and Notch1 is depicted in this diagram. (F) The effect of ZFAS1 on Notch1 promoter activity in HEK293T cells with the use of a dual-luciferase reporter incorporating the Notch promoter. n = 3. ∗∗P < 0.01 vs Ctl. (G) The interaction of Notch1 with ZFAS1 in cardiomyocytes was examined with the use of the ChiRP assay. n = 3. ∗∗P < 0.01 vs AS-ZFAS1. (H) RIP analysis of specific associations of DNMT3b, DNMT3a, and DNMT1 with ZFAS1. n = 3. ∗P < 0.05 vs IgG; ∗∗P < 0.01 vs IgG. (I) ChIP analysis of associations of DNMT3b, DNMT3a, and DNMT1 with Notch1. n = 3. ∗P < 0.05 vs IgG. P values were determined by means of t-tests and 1-way analyses of variance with Bonferroni multiple group comparisons. Data are presented as mean ± SEM. DNMT = DNA methyltransferase; PCR = polymerase chain reaction; other abbreviations as in Figures 1 and 2.